













| General Information: |
|
| Name(s) found: |
FMT1 /
YBL013W
[SGD]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 401 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrion
[IDA |
| Biological Process: |
methionyl-tRNA aminoacylation
[IDA translational initiation [IDA |
| Molecular Function: |
methionyl-tRNA formyltransferase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVKMRRITPT RLLFTCRYIS NNASPPVQPL NVLFFGSDTF SNFSLQALNE LRQNNGSCGI 60
61 VDNIQVVTRS PKWCGRQKSI LKYPPIFDMA EKLQLPRPIT CDTKQEMLAL SKLTPSRQGN 120
121 PENDGSGAPF NAIIAVSFGK LIPGDLIRAV PLALNVHPSL LPRHKGSAPI QRALLEGDTY 180
181 TGVTIQTLHP DRFDHGAIVA QTEPLAIATM LSKGRVNDST ADFNSEGLPR RTAILMDQLG 240
241 ALGAQLLGQT LRERLYLPQN RVQAPTAYKP SYAHRITTED KRIHWARDSA AELLNKLETL 300
301 GPLHAFKEAT AARKDAQNSV LKRILFHECK VMRDARLDNG SKPGMFKYDD IKDCILVTCR 360
361 GNLLLCVSRL QFEGFAVERA GQFMARLRKR CGALSEKLVF L |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
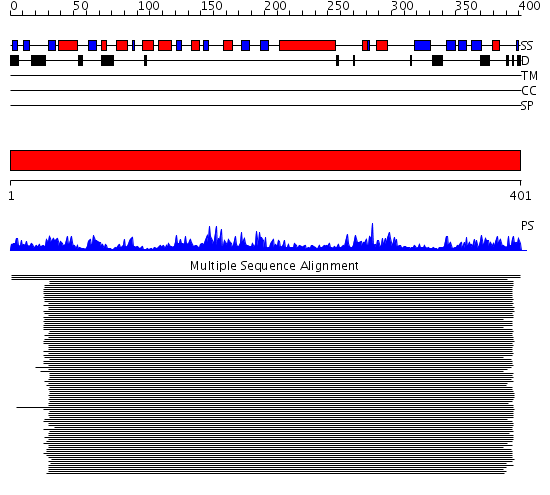
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..401] | 75.0 | Methionyl-tRNAfmet formyltransferase, C-terminal domain; Methionyl-tRNAfmet formyltransferase |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.83 |
Source: Reynolds et al. (2008)