













| General Information: |
|
| Name(s) found: |
HEXO2_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 11 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 580 amino acids |
Gene Ontology: |
|
| Cellular Component: |
plasma membrane
[IDA]
|
| Biological Process: |
carbohydrate metabolic process
[IEA]
|
| Molecular Function: |
hexosaminidase activity
[IDA]
UDP-glucosyltransferase activity [IDA] hydrolase activity, hydrolyzing O-glycosyl compounds [ISS] beta-N-acetylhexosaminidase activity [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLTLSKFHVI LIPILFFITL LSPLFSIALP INIWPKPRFL SWPQHKAIAL SPNFTILAPE 60
61 HQYLSASVTR YHNLIRSENY SPLISYPVKL MKRYTLRNLV VTVTDFSLPL HHGVDESYKL 120
121 SIPIGSFSAH LLAHSAWGAM RGLETFSQMI WGTSPDLCLP VGIYIQDSPL FGHRGVLLDT 180
181 SRNYYGVDDI MRTIKAMSAN KLNVFHWHIT DSQSFPLVLP SEPSLAAKGS LGPDMVYTPE 240
241 DVSKIVQYGF EHGVRVLPEI DTPGHTGSWG EAYPEIVTCA NMFWWPAGKS WEERLASEPG 300
301 TGQLNPLSPK TYEVVKNVIQ DIVNQFPESF FHGGGDEVIP GCWKTDPAIN SFLSSGGTLS 360
361 QLLEKYINST LPYIVSQNRT VVYWEDVLLD AQIKADPSVL PKEHTILQTW NNGPENTKRI 420
421 VAAGYRVIVS SSEFYYLDCG HGGFLGNDSI YDQKESGGGS WCAPFKTWQS IYNYDIADGL 480
481 LNEEERKLVL GGEVALWSEQ ADSTVLDSRL WPRASALAES LWSGNRDERG VKRCGEAVDR 540
541 LNLWRYRMVK RGIGAEPIQP LWCLKNPGMC NTVHGALQDQ |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
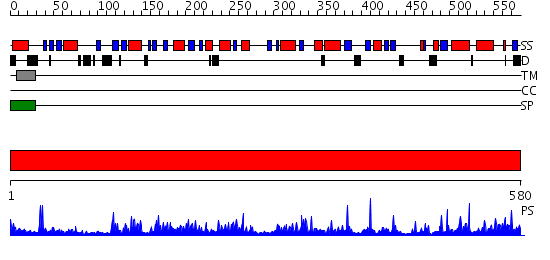
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..580] | 145.0 | Human beta-Hexosaminidase B |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Source: Reynolds et al. 2008. Manuscript submitted |

|