













| General Information: |
|
| Name(s) found: |
wwp-1 /
CE31384
[WormBase]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 794 amino acids |
Gene Ontology: |
|
| Cellular Component: |
intracellular
[IEA |
| Biological Process: |
embryonic development ending in birth or egg hatching
[IMP protein modification process [IEA ubiquitin cycle [IEA |
| Molecular Function: |
ubiquitin-protein ligase activity
[IEA protein binding [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MARNEPSSQQ PSSSGSNGTP AQQNGSAKPS KVTVKVVNAS FTKAADCYVE ITSDTSSAAP 60
61 KKTTVKKKTM APEWNEHLNV HANESSTISF RLLQKAKLFD DTCLGMAKLK LSSLTRNENG 120
121 EFKNDINNIS LLAKDSSKIG TLNIIFSGYP ERKRRSAGVR AETAASASSE ASTSNGVATS 180
181 SSARRPATAK RDTLAAPTST AAAAAAATAG GTPAAGAEEQ LPDGWEMRFD QYGRKYYVDH 240
241 TTKSTTWERP STQPLPQGWE MRRDPRGRVY YVDHNTRTTT WQRPTADMLE AHEQWQSGRD 300
301 QAMLQWEQRF LLQQNNFSAD DPLGPLPEGW EKRQDPNTSR MYFVNHVNRT TQWEDPRTQG 360
361 FRGSDQPLPD GWEMRFTEQG VPFFIDHQSK TTTYNDPRTG KPVGPLGVVG VQMAMEKSFR 420
421 WKIAQFRYLC LSNSVPNHVK ITVSRNNVFE DSFQEIMRKN AVDLRRRLYI QFRGEEGLDY 480
481 GGVAREWFFL LSHEVLNPMY CLFMYAGNNN YSLQINPASF VNPDHLKYFE YIGRFIAMAL 540
541 FHGKFIYSGF TMPFYKKMLN KKIVLKDIEQ VDSEIYNSLM WIKDNNIDEC DMELYFVADY 600
601 ELLGELKTYE LKEGGTEIAV TEENKLEYIE LLVEWRFNRG VEQQTKAFFT GFNSVFPLEW 660
661 MQYFDERELE LLLCGMQDVD VDDWQRNTVY RHYAPQSKQV TWFWQWVRSL DQEKRARLLQ 720
721 FVTGTCRVPV GGFSELMGST GPQLFCIERV GKENWLPRSH TCFNRLDLPP YRSYDQLVEK 780
781 LSMAIEMTEG FGNE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
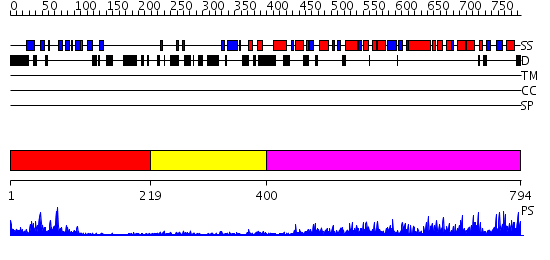
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..218] | 24.39794 | Synaptogamin I | |
| 2 | View Details | [219..399] | 3.32 | NMR structure of WW domains (WW3-4) from Suppressor of Deltex | |
| 3 | View Details | [400..794] | 119.0 | Conformational Flexibility Underlies Ubiquitin Ligation Mediated by the WWP1 HECT domain E3 Ligase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.91 |
Source: Reynolds et al. (2008)