













| General Information: |
|
| Name(s) found: |
pdr-1 /
CE31713
[WormBase]
|
| Description(s) found:
Found 15 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 386 amino acids |
Gene Ontology: |
|
| Cellular Component: |
ubiquitin ligase complex
[IPI]
cytoplasm [IDA] neuronal cell body [IDA] |
| Biological Process: |
embryonic cleavage
[IMP embryonic development ending in birth or egg hatching [IMP protein modification process [IEA cytokinesis [IMP |
| Molecular Function: |
zinc ion binding
[IEA protein binding [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSDEISILIQ DRKTGQRRNL TLNINITGNI EDLTKDVEKL TEIPSDELEV VFCGKKLSKS 60
61 TIMRDLSLTP ATQIMLLRPK FNSHNENGAT TAKITTDSSI LGSFYVWCKN CDDVKRGKLR 120
121 VYCQKCSSTS VLVKSEPQNW SDVLKSKRIP AVCEECCTPG LFAEFKFKCL ACNDPAAALT 180
181 HVRGNWQMTE CCVCDGKEKV IFDLGCNHIT CQFCFRDYLL SQLERFGFVN QPPHGFTIFC 240
241 PYPGCNRVVQ DVHHFHIMGQ TSYSEYQRKA TERLIAVDDK GVTCPNVSCG QSFFWEPYDD 300
301 DGRSQCPDCF FSFCRKCFER NCVCQSEDDL TRTTIDATTR RCPKCHVATE RNGGCAHIHC 360
361 TSCGMDWCFK CKTEWKEECQ WDHWFN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
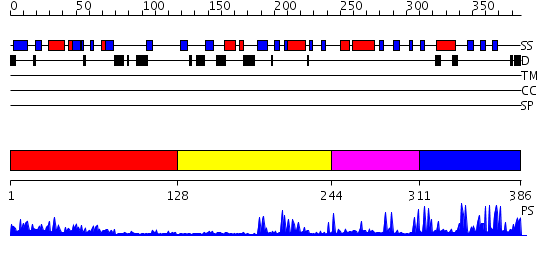
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..127] | 16.30103 | Structure of the DNA repair protein hHR23a | |
| 2 | View Details | [128..243] | 5.0 | Solution Structure of the RING finger Domain of the human UbcM4-interacting Protein 4 | |
| 3 | View Details | [244..310] | 1.3 | No description for 2jmoA was found. | |
| 4 | View Details | [311..386] | 8.09691 | Solution Structure of the C-terminal RING from a RING-IBR-RING (TRIAD) motif |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)