













| General Information: |
|
| Name(s) found: |
klp-10 /
CE04148
[WormBase]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 690 amino acids |
Gene Ontology: |
|
| Cellular Component: |
microtubule associated complex
[IEA |
| Biological Process: |
embryonic cleavage
[IMP embryonic development ending in birth or egg hatching [IMP cytokinesis [IMP chromosome segregation [IMP microtubule-based movement [IEA |
| Molecular Function: |
ATP binding
[IEA microtubule motor activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSQYIKCYGR VRPSARLGAS SESLVVGGRV VTIDTDNGSK RYELHHIFDS SSSQEDVFGT 60
61 VAKKIVEDCV EGYNGTVFAY GQTGSGKTHT MLGPCDSWTD PELMGLIPRS VEHVFQHLDT 120
121 KAKKCQKLTF SVSVEFVELY NEVIYDLLNA KNKVQLRDSG KDIQLVGALP KNVDNPLDLM 180
181 HLVQKGWQER STGSTAMNAE SHALLIIRIK TQERTGELVK ERSSILNLVD LAGSERQTHT 240
241 KSSGDRLKEA TNINSSLTVL GRCIRLLADP SKAKGHVPYR DSHLTHILKN SLGGNSKTAV 300
301 IVNMHPDRDF AQESNSTLMF AQSCTMIMNI ATRNEVMTGD QESSYKKAIQ QLRQEKRGKS 360
361 TANSSSTFEC IALKNQTLQL EIDASEKRYQ ELQKETSALR NRYQENLDAT ILMQTPSAKE 420
421 RRSSSRPKRR ETQYRPSPAR MALFAEVETS EDAVLDLQCQ NEKLCCELSS LKDKHEEATS 480
481 KSTETERLLQ GEIVALQKEV SIVSESLRKS EAVAEGYKEK VAANNLTLDK LLLHIDASRK 540
541 RIAELELSSK ESENRSSEEL EVDELQLSNA TIKKLNADLS TKAGLIRSLE DSIEKKSQMI 600
601 TRLEEQAVLD SSELSSTKRQ LEHSRLEAIQ WRKISDETTS AMQTQLKDHQ TDIDQCRHKK 660
661 KSEIQSMQLS LDNANARVKE SEAMMKKTSG |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
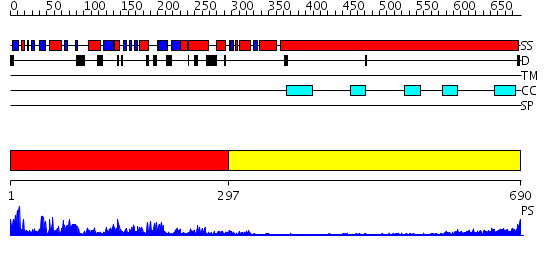
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..296] | 85.39794 | Kinesin | |
| 2 | View Details | [297..690] | 12.0 | Heavy meromyosin subfragment |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)