













| General Information: |
|
| Name(s) found: |
FBpp0307743
[FlyBase]
Apc-PA / FBpp0084720 [FlyBase] |
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 2417 amino acids |
Gene Ontology: |
|
| Cellular Component: |
apical part of cell
[IDA]
cytoplasm [IDA] microtubule associated complex [ISS] axon [IDA] |
| Biological Process: |
cell-cell adhesion
[IGI]
chitin-based larval cuticle pattern formation [IGI] head involution [IGI] axon guidance [IGI] brain development [IGI] imaginal disc pattern formation [IGI] microtubule-based process [ISS] neuroblast proliferation [IGI] medulla oblongata development [IGI] negative regulation of compound eye retinal cell programmed cell death [IMP] negative regulation of Wnt receptor signaling pathway [IGI] regulation of Wnt receptor signaling pathway [IGI] oocyte localization during germarium-derived egg chamber formation [IGI] |
| Molecular Function: |
beta-catenin binding
[IDA]
microtubule binding [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLEPTLTPDL EEGIAELSLE DVDVDSVPER KPHFLDYDAV PPPDYEEGFG ASSGEQLKMD 60
61 KKYERDGEVS DYELAASGYT KKEFTQDDNT LHFTQSAVGL GSGAVGKKPA AKHFLDENPI 120
121 PPDYMLAQEL REMREHRSLD RNFERQSAQQ QQLDELPPRN GGGSPASAGR PSRSKEPSYT 180
181 LSRFLDGDAP APAPRLPKGA AWTTSFDERY TSSAVEATLG SKVECVYSLL SMLGSNDPLE 240
241 MAKKFLELSG NAQSCATLRR SGCMPLLVQM MHAPDNDQEV RKCAGQALHN VVHSHPDEKA 300
301 GRREAKVLRL LDQIVDYCSF LKTLLQSGGE AIADDSDRHP LAAISSLMKV SFDEEHRHAM 360
361 CELGALHAIP NLVHLDHAVH GPKPEDQCCN SLRRYALMAL TNLTFGDENN KALLCGQKQF 420
421 MEALVAQLDS APDDLLQVTA SVLRNLSWRA DSNMKAVLNE IGTVTALALA AMRNRSENTL 480
481 KAILSALWNL SAHCSTNKAE FCAVDGALAF LVGMLSYEGP SKTLKIIENA GGILRNVSSH 540
541 IAVCEPYRQI LRQHNCLAIL LQQLKSESLT VVSNSCGTLW NLSARSAEDQ KFLWDNGAVP 600
601 MLRSLIHSKH AMISEGSSSA LKNLLNFRPA VQNHHQLDPI ARSMGLKALP TLEARKAKAL 660
661 QQELGERHTA ETCDNLDTGG KLDKERASSS SRRHPAPRLT RSAMLTKSES RDSVYSAKSD 720
721 CAYDHLIRSA SASDAHRKVK PKITDFDLEM EQDTEATEEQ PIDYSVKYSE NATKTSTYQE 780
781 TDLDQPTDFS LRYAENQIES DLDISGPAGG QKSTITPPAE TVPEKSEGQE ILLILDDSVK 840
841 CYQTEDTPYV ISNAASVTDL RVAAKADAEA EVKPEVREVT SKEGAPKKLP KLSQCGSGSY 900
901 TPEKPINYCE EGTPGYFSRY DSLSSLDESG KANQAIVGTD ADIKPKLEKQ EEQESQPAEQ 960
961 VLTKPPTQAN SALETPLMFS RRSSMDSLVH DPDVDVANCD DKSSVVSDFS RLASGVISPS1020
1021 EIPDSPTQSM PQSPRRNSVA GSGQNVDSPP VVIPASLQPL RSVFEDDLSS FNVEHTPAQF1080
1081 STATSLSNLS IVDDEKAPAS VAEEDNEDEL LLANCINMGM QRKPTEAVKS TVVNSEVDVA1140
1141 EETIRSYCTE DTPALLSKVP SNTNLSVISM SSTDPKDATA GQAQMYAHQL SDDVSSNASD1200
1201 CGGASGHLLQ QCIRDGMKKP LGEATSDPIA MLRRGGNELP GYLPSADEMN KFLVEDSPCN1260
1261 FSVVSGLSNL TVGSSLVGPA VQLKETEPSS ADQNPEMKAK PGKQEQVRRP PHWQDDSLSS1320
1321 LSIDSEDDTN LLSQAIAAGC NRPKSNLGFS SNGKRSSSLS SSQPIAINAA TSASSLNSAM1380
1381 TVRKSQQQES YSSVDSSDSN DNQSKSLFEL CILKGMYKTK EPGARAQQMQ EQPIVGSSSV1440
1441 QSNPSLKQFD SLPVQLPSSG QVKRQRHHHH HHHHRERERE RKDEKLLQEC INTGISKKIN1500
1501 AVPKNVLATS AAALEPCHPM AATTSASALS TAAPDVEQKA HATSNPQQQS STHPSSHILP1560
1561 NPIDAIATVT DTVRSPAAPN QGNGNASQNG LETATGSKDL DSEDRSSDES NQSFIMETMV1620
1621 RLDSALNETC ISGASEKHKD PDLMLKSVER LTMEFVTSAE QLRSSSHNHS SSNSHKNNSS1680
1681 NNTWNESTCP NDVSFPSVSQ TAPVLASLSL DEDATEARSL HELIEITPTN EQQPESLEGE1740
1741 TDTLVNGHAD SYSGSSGGLN FQLGGQVQNA GVRLEPQRLL FNGTSASIMT NSTMIAFEAR1800
1801 ALAENLLQPA ATDDDTTEMT FSLNSLDLDN IRPPSGMESL NSCYQDHSQP SSLRQAMPSK1860
1861 SPRFARKMFP ANLVARRALG HLAGSAESVN SSCNLLDNIK PPSLMDELLD SMISVDSIQS1920
1921 EVADGEQDCS MATTISVSNY ETAACDDQTM TVLQSCFDED EDATMNDYSS AESTPKHGST1980
1981 PSPNRRSLTP KDKRRLTKDR FKTYTIATSC EMEAPEANET LQIEIVEAAV PVATPSPRAN2040
2041 GRRRGSAERY KTQLIECPLA LIQPQPDDCP SEQLSSIRAM MQQFTFITDI NIGHSQETCE2100
2101 STDHPEDAGE SPECDQNSET ESCDGQEPDQ LPPPPSIVDL RTSVVKPTTL EPATAVKLVR2160
2161 GRKKPAYVSP YSMQSQRNSN NAAPSKKKTL SPTIAKRSLV PGGSGVRLPA KKKPTPPPEP2220
2221 APARLERQGT FVKDEPTNSN VQVPVVETKP AQTSPTHRAS KLPTKKGTAS GGSPSKAGSP2280
2281 KRIPLAPARR MTPQRANTSL RLAAGKSHAA SRVVSGRVSS TTPPSRSNSN LNGSSAAAAA2340
2341 AAKINHAQSR IANIWKRVDE AKTKQSSSNL RTQKTKSSNM LNANGTKPTL LRSSTFDNTP2400
2401 STAGGVKSKL PVVGARK |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
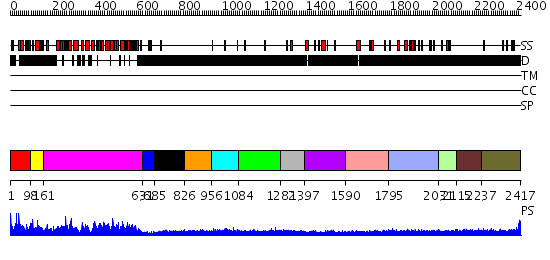
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..97] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [98..160] | 14.69897 | Tumor suppressor gene product Apc | |
| 3 | View Details | [161..630] | 126.0 | beta-Catenin | |
| 4 | View Details | [631..684] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [685..825] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [826..955] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [956..1083] | 2.45 | Beta-catenin in complex with a phosphorylated APC 20aa repeat fragment | |
| 8 | View Details | [1084..1281] | N/A | No confident structure predictions are available. | |
| 9 | View Details | [1282..1396] | 1.132988 | View MSA. No confident structure predictions are available. | |
| 10 | View Details | [1397..1589] | N/A | No confident structure predictions are available. | |
| 11 | View Details | [1590..1794] | N/A | No confident structure predictions are available. | |
| 12 | View Details | [1795..2030] | N/A | No confident structure predictions are available. | |
| 13 | View Details | [2031..2114] | N/A | No confident structure predictions are available. | |
| 14 | View Details | [2115..2236] | 1.128988 | View MSA. No confident structure predictions are available. | |
| 15 | View Details | [2237..2417] | 1.117985 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 8 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 9 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 10 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 11 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 12 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 13 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 14 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 15 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)