













| General Information: |
|
| Name(s) found: |
C10orf2
[HGNC (HUGO)]
|
| Description(s) found:
Found 38 descriptions. SHOW ALL |
|
| Organism: | Homo sapiens |
| Length: | 684 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MWVLLRSGYP LRILLPLRGE WMGRRGLPRN LAPGPPRRRY RKETLQALDM PVLPVTATEI 60
61 RQYLRGHGIP FQDGHSCLRA LSPFAESSQL KGQTGVTTSF SLFIDKTTGH FLCMTSLAEG 120
121 SWEDFQASVE GRGDGAREGF LLSKAPEFED SEEVRRIWNR AIPLWELPDQ EEVQLADTMF 180
181 GLTKVTDDTL KRFSVRYLRP ARSLVFPWFS PGGSGLRGLK LLEAKCQGDG VSYEETTIPR 240
241 PSAYHNLFGL PLISRRDAEV VLTSRELDSL ALNQSTGLPT LTLPRGTTCL PPALLPYLEQ 300
301 FRRIVFWLGD DLRSWEAAKL FARKLNPKRC FLVRPGDQQP RPLEALNGGF NLSRILRTAL 360
361 PAWHKSIVSF RQLREEVLGE LSNVEQAAGL RWSRFPDLNR ILKGHRKGEL TVFTGPTGSG 420
421 KTTFISEYAL DLCSQGVNTL WGSFEISNVR LARVMLTQFA EGRLEDQLDK YDHWADRFED 480
481 LPLYFMTFHG QQSIRTVIDT MQHAVYVYDI CHVIIDNLQF MMGHEQLSTD RIAAQDYIIG 540
541 VFRKFATDNN CHVTLVIHPR KEDDDKELQT ASIFGSAKAS QEADNVLILQ DRKLVTGPGK 600
601 RYLQVSKNRF DGDVGVFPLE FNKNSLTFSI PPKNKARLKK IKDDTGPVAK KPSSGKKGAT 660
661 TQNSEICSGQ APTPDQPDTS KRSK |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Cheeseman, I.M., et al. (2004) | |
| View Run | No Comments | Cheeseman, I.M., et al. (2004) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
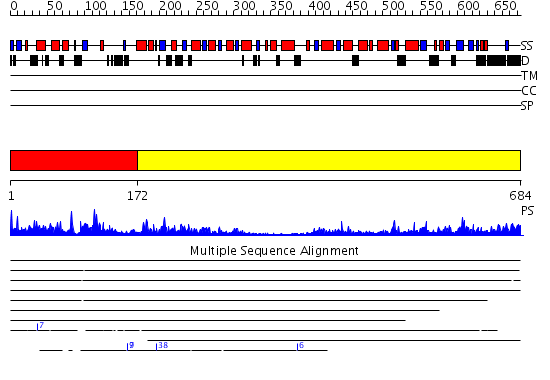
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..171] | 50.69897 | Crystal Structure of the Aquifex aeolicus primase (Zinc Binding and RNA Polymerase Domains) | |
| 2 | View Details | [172..684] | 51.522879 | The Crystal Structure of the Bifunctional Primase-Helicase of Bacteriophage T7 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)