













| General Information: |
|
| Name(s) found: |
YPK1 /
YKL126W
[SGD]
|
| Description(s) found:
Found 23 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 680 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular bud neck
[IDA cytosol [IDA plasma membrane [IDA |
| Biological Process: |
protein amino acid phosphorylation
[IMP endocytosis [IMP sphingolipid metabolic process [IMP |
| Molecular Function: |
protein serine/threonine kinase activity
[IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MYSWKSKFKF GKSKEEKEAK HSGFFHSSKK EEQQNNQATA GEHDASITRS SLDRKGTINP 60
61 SNSSVVPVRV SYDASSSTST VRDSNGGNSE NTNSSQNLDE TANIGSTGTP NDATSSSGMM 120
121 TIKVYNGDDF ILPFPITSSE QILNKLLASG VPPPHKEISK EVDALIAQLS RVQIKNQGPA 180
181 DEDLISSESA AKFIPSTIML PGSSTLNPLL YFTIEFDNTV ATIEAEYGTI AKPGFNKIST 240
241 FDVTRKLPYL KIDVFARIPS ILLPSKTWQQ EMGLQDEKLQ TIFDKINSNQ DIHLDSFHLP 300
301 INLSFDSAAS IRLYNHHWIT LDNGLGKINI SIDYKPSRNK PLSIDDFDLL KVIGKGSFGK 360
361 VMQVRKKDTQ KVYALKAIRK SYIVSKSEVT HTLAERTVLA RVDCPFIVPL KFSFQSPEKL 420
421 YFVLAFINGG ELFYHLQKEG RFDLSRARFY TAELLCALDN LHKLDVVYRD LKPENILLDY 480
481 QGHIALCDFG LCKLNMKDDD KTDTFCGTPE YLAPELLLGL GYTKAVDWWT LGVLLYEMLT 540
541 GLPPYYDEDV PKMYKKILQE PLVFPDGFDR DAKDLLIGLL SRDPTRRLGY NGADEIRNHP 600
601 FFSQLSWKRL LMKGYIPPYK PAVSNSMDTS NFDEEFTREK PIDSVVDEYL SESVQKQFGG 660
661 WTYVGNEQLG SSMVQGRSIR |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |
|
TEC1 YPK1 YPK2 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
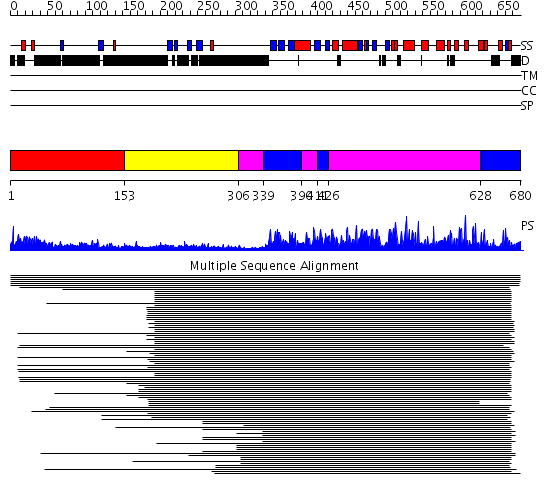
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..152] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [153..305] | 18.0 | C2 domain from protein kinase c (beta) | |
| 3 | View Details | [306..338] [390..410] [426..627] |
890.0 | cAMP-dependent PK, catalytic subunit | |
| 4 | View Details | [339..389] [411..425] [628..680] |
890.0 | cAMP-dependent PK, catalytic subunit |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)