













| General Information: |
|
| Name(s) found: |
RGT1 /
YKL038W
[SGD]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1170 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IC |
| Biological Process: |
negative regulation of transcription
[IDA regulation of glucose import [IEP glucose metabolic process [TAS |
| Molecular Function: |
DNA binding
[IDA RNA polymerase II transcription factor activity [TAS transcription corepressor activity [TAS transcription activator activity [TAS transcription repressor activity [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNELNTVSTN SSDSTKNGGT SNSPDDMDSA AAASHAIKKR TKASRACDQC RKKKIKCDYK 60
61 DEKGVCSNCQ RNGDRCSFDR VPLKRGPSKG YTRSTSHPRT NEIQDHNNSR SYNTFDNSNN 120
121 TLNNNTGNSG DNGINSNTVP STPSRSNSVL LPPLTQYIPQ AGGIPPSFQN PAIQSTMPAG 180
181 NIGQQQFWKV PYHEFQHQRK GSIDSLQSDI SVRTLNPNEQ LSYNTVQQSP ITNKHTNDSG 240
241 NANGSVTGSG SASGSGGYWS FIRTSGLLAP TDDHNGEQTR RSSSIPSLLR NTSNSLLLGG 300
301 QPQLPPPQQQ SQPQAHQQKL QQGQNLYSYS QFSQQQPYNP SISSFGQFAA NGFHSRQGSV 360
361 ASEAMSPSAP AMFTSTSTNP VNVAQQTQRP QGQQVPQFSS ELDGNKRRQS APVSVTLSTD 420
421 RLNGNENNNG EINNNNGSNN SGSSKDTSQH SQESVTTPAA LEASSPGSTP QRSTKKRRKS 480
481 YVSKKTKPKR DSSISITSKD SAHPMTTSST IAYGQISDVD LIDTYYEFIH VGFPIIPLNK 540
541 TTLTSDLLLV NTQPISNIHE VNSYVILWFR NSLELLVRVA LKQKPGGKFF DNIVGVALSP 600
601 SNDNNKAGFT TATARDDAEK TRRDSHNEVQ DTLEVQSVFI AALNECFQKI VDIHPKFREN 660
661 NDQISPKIKV IYLSTFILLN YILAFVGYDN SFVLGMSVTI FNEFKLYKLL LFPEPDINDV 720
721 KPPVDEEVST GNGNTKTSEF EIGSESAGHM NPSNSPNSMD ENISHYSVLF KRLYVLLSVF 780
781 DSLQSCAFGG PKLLNISIQG STERFFSNDL GSKWCLEQSQ LRLKSVLQSL KLGELMSELT 840
841 RNRISMNGNR KPGFDITNSS SLLSEYVETQ PLSVAQLFCK LLIGKHNFIN CLLSLYDSEA 900
901 GVYSDLTLDL SSKIADSLCS LISIILQVLT LILRLNPTNS IDFNYRPPNP PANNPTVQEG 960
961 PSAMGSSPVA GNLSAAPPSE GNPDFYKKLL GLKQDTGTIL SDLCRGIISP FAIAILHEVY1020
1021 NITELVKQMP TSLISIMMTA TTTQNTQDTK KSQDLVMKLS NSMNEVVQIT SVLTMIKPFK1080
1081 IFEHELNKPI MSLTGGLSST TRNDVMWPKS GQGLRESSVM KTLLDERRTS GTQPTTAPVA1140
1141 AEEPRLENVA LENFVSIGWK LLDDSELGWY |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample bir 1 from feb 2005 | Sandall S, et all (2006) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #25 Asynchronous Prep3-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
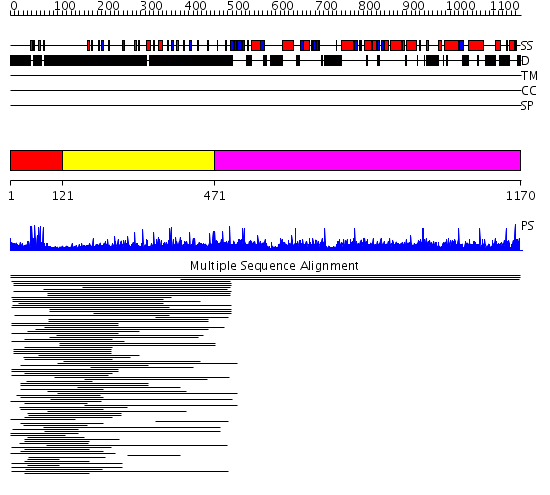
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..120] | 10.0 | Hap1 (Cyp1); HAP1 | |
| 2 | View Details | [121..470] | 28.369997 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [471..1170] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [458..602] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [603..722] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [723..976] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [977..1170] | 1.005945 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||
| 6 | No functions predicted. | ||||||||||||||||||||||||||||||
| 7 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.90 |
Source: Reynolds et al. (2008)