













| General Information: |
|
| Name(s) found: |
SLN1 /
YIL147C
[SGD]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1220 amino acids |
Gene Ontology: |
|
| Cellular Component: |
plasma membrane
[ISS |
| Biological Process: |
osmosensory signaling pathway via two-component system
[IDA response to hydrogen peroxide [IMP protein amino acid phosphorylation [TAS |
| Molecular Function: |
two-component sensor activity
[TAS osmosensor activity [IDA protein histidine kinase activity [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MRFGLPSKLE LTPPFRIGIR TQLTALVSIV ALGSLIILAV TTGVYFTSNY KNLRSDRLYI 60
61 AAQLKSSQID QTLNYLYYQA YYLASRDALQ SSLTSYVAGN KSADNWVDSL SVIQKFLSSS 120
121 NLFYVAKVYD SSFNAVLNAT NNGTGDLIPE DVLDSLFPLS TDTPLPSSLE TIGILTDPVL 180
181 NSTDYLMSMS LPIFANPSII LTDSRVYGYI TIIMSAEGLK SVFNDTTALE HSTIAIISAV 240
241 YNSQGKASGY HFVFPPYGSR SDLPQKVFSI KNDTFISSAF RNGKGGSLKQ TNILSTRNTA 300
301 LGYSPCSFNL VNWVAIVSQP ESVFLSPATK LAKIITGTVI AIGVFVILLT LPLAHWAVQP 360
361 IVRLQKATEL ITEGRGLRPS TPRTISRASS FKRGFSSGFA VPSSLLQFNT AEAGSTTSVS 420
421 GHGGSGHGSG AAFSANSSMK SAINLGNEKM SPPEEENKIP NNHTDAKISM DGSLNHDLLG 480
481 PHSLRHNDTD RSSNRSHILT TSANLTEARL PDYRRLFSDE LSDLTETFNT MTDALDQHYA 540
541 LLEERVRART KQLEAAKIEA EAANEAKTVF IANISHELRT PLNGILGMTA ISMEETDVNK 600
601 IRNSLKLIFR SGELLLHILT ELLTFSKNVL QRTKLEKRDF CITDVALQIK SIFGKVAKDQ 660
661 RVRLSISLFP NLIRTMVLWG DSNRIIQIVM NLVSNALKFT PVDGTVDVRM KLLGEYDKEL 720
721 SEKKQYKEVY IKKGTEVTEN LETTDKYDLP TLSNHRKSVD LESSATSLGS NRDTSTIQEE 780
781 ITKRNTVANE SIYKKVNDRE KASNDDVSSI VSTTTSSYDN AIFNSQFNKA PGSDDEEGGN 840
841 LGRPIENPKT WVISIEVEDT GPGIDPSLQE SVFHPFVQGD QTLSRQYGGT GLGLSICRQL 900
901 ANMMHGTMKL ESKVGVGSKF TFTLPLNQTK EISFADMEFP FEDEFNPESR KNRRVKFSVA 960
961 KSIKSRQSTS SVATPATNRS SLTNDVLPEV RSKGKHETKD VGNPNMGREE KNDNGGLEQL1020
1021 QEKNIKPSIC LTGAEVNEQN SLSSKHRSRH EGLGSVNLDR PFLQSTGTAT SSRNIPTVKD1080
1081 DDKNETSVKI LVVEDNHVNQ EVIKRMLNLE GIENIELACD GQEAFDKVKE LTSKGENYNM1140
1141 IFMDVQMPKV DGLLSTKMIR RDLGYTSPIV ALTAFADDSN IKECLESGMN GFLSKPIKRP1200
1201 KLKTILTEFC AAYQGKKNNK |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |
|
ARF2 COX2 MMF1 SLN1 YDR018C YDR374C |
| View Details | Ho Y, et al. (2002) |

|
COP1 SLN1 |
| View Details | Qiu et al. (2008) |

|
MID2 MSB2 SHO1 SLN1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #19 Asynchronous Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #30 Asynchronous Prep6-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) | |
| View Run | #19b Asynchronous Prep1-new search criteria | Keck JM, et al. (2011) | |
| View Run | #19a Asynchronous Prep1-new search criteria | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
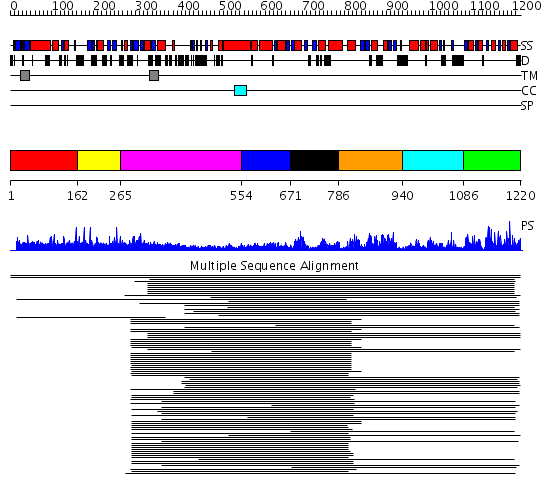
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..161] | 1.004953 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [162..264] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [265..553] | 3.553989 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [554..670] | 14.04 | EnvZ histidine kinase | |
| 5 | View Details | [671..785] | 4.272994 | View MSA. Confident ab initio structure predictions are available. | |
| 6 | View Details | [786..939] | 81.68867 | Histidine kinase domain of the osmosensor EnvZ | |
| 7 | View Details | [940..1085] | 13.69897 | Transcriptional regulatory protein DctD, receiver domain | |
| 8 | View Details | [1086..1220] | 123.156545 | CheY protein |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 8 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|