













| General Information: |
|
| Name(s) found: |
YEN1 /
YER041W
[SGD]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 759 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[ISS |
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
single-stranded DNA specific endodeoxyribonuclease activity
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGVSQIWEFL KPYLQDSRIP LRKFVIDFNK SQKRAPRIAI DAYGWLFECG FIQNIDISPR 60
61 SRSRSRSPTR SPRDSDIDSS QEYYGSRSYT TTGKAVINFI SRLKELLSLN VEFLLVFDGV 120
121 MKPSFKRKFN HEQNATTCDD EKEYYSSWEQ HVKNHEVYGN CKGLLAPSDP EFISLVRKLL 180
181 DLMNISYVIA CGEGEAQCVW LQVSGAVDFI LSNDSDTLVF GGEKILKNYS KFYDDFGPSS 240
241 ITSHSPSRHH DSKESFVTVI DLPKINKVAG KKFDRLSLLF FSVLLGADYN RGVKGLGKNK 300
301 SLQLAQCEDP NFSMEFYDIF KDFNLEDLTS ESLRKSRYRL FQKRLYLYCK DHSVELFGRN 360
361 YPVLLNQGSF EGWPSTVAIM HYFHPIVQPY FDEEVLSDKY INMAGNGHYR NLNFNELKYF 420
421 LQSLNLPQIS SFDKWFHDSM HEMFLLREFL SIDESDNIGK GNMRITEEKI MNIDGGKFQI 480
481 PCFKIRYTTF LPNIPISSQS PLKRSNSPSR SKSPTRRQMD IMEHPNSLWL PKYLIPQSHP 540
541 LVIQYYETQQ LIQKEKEKKG KKSNKSRLPQ KNNLDEFLRK HTSPIKSIGK VGESRKEILE 600
601 PVRKRLFVDT DEDTSLEEIP APTRLTTVDE HSDNDDDSLI FVDEITNSQS VLDSSPGKRI 660
661 RDLTQDEQVD VWKDVIEISP IKKSRTTNAE KNPPESGLKS RSSITINARL QGTKMLPPNL 720
721 TAPRLEREHS SVLDQLVTDA QDTVDRFVAC DSDSSSTIE |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |

|
CSI1 CSN9 ERV46 FPR2 HUL5 IQG1 MOT1 MPT5 NAS6 NMD3 NPT1 PCI8 PEP1 PST1 PUS9 RPN1 RPN10 RPN11 RPN12 RPN13 RPN14 RPN2 RPN5 RPN6 RPN7 RPN8 RPN9 RPT1 RPT2 RPT3 RPT4 RPT5 RPT6 RRI1 RRI2 SEC18 SEM1 SKP1 SPG5 SPO77 SQT1 SWA2 TCB2 TIP41 TOD6 TOR2 TSR4 UBP6 YEN1 YKL027W YMR258C YNL022C YPR003C YPT52 |
| View Details | Ho Y, et al. (2002) |
|
FPR4 POL30 RPF2 YEN1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #25 Asynchronous Prep3-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Lockshon D, et al. (2006) |
New Feature: Upload Your Own Microscopy Data
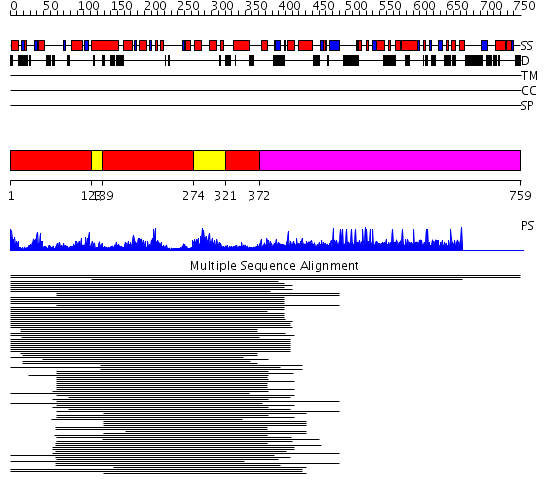
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..122] [139..273] [321..371] |
133.512579 | Flap endonuclease-1 (Fen-1 nuclease) | |
| 2 | View Details | [123..138] [274..320] |
133.512579 | Flap endonuclease-1 (Fen-1 nuclease) | |
| 3 | View Details | [372..759] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.92 |
Source: Reynolds et al. (2008)