













| General Information: |
|
| Name(s) found: |
YND1 /
YER005W
[SGD]
|
| Description(s) found:
Found 24 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 630 amino acids |
Gene Ontology: |
|
| Cellular Component: |
Golgi apparatus
[IDA COPI-coated vesicle [IDA |
| Biological Process: |
protein amino acid glycosylation
[IMP |
| Molecular Function: |
nucleoside-diphosphatase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLIENTNDRF GIVIDAGSSG SRIHVFKWQD TESLLHATNQ DSQSILQSVP HIHQEKDWTF 60
61 KLNPGLSSFE KKPQDAYKSH IKPLLDFAKN IIPESHWSSC PVFIQATAGM RLLPQDIQSS 120
121 ILDGLCQGLK HPAEFLVEDC SAQIQVIDGE TEGLYGWLGL NYLYGHFNDY NPEVSDHFTF 180
181 GFMDMGGAST QIAFAPHDSG EIARHRDDIA TIFLRSVNGD LQKWDVFVST WLGFGANQAR 240
241 RRYLAQLINT LPENTNDYEN DDFSTRNLND PCMPRGSSTD FEFKDTIFHI AGSGNYEQCT 300
301 KSIYPLLLKN MPCDDEPCLF NGVHAPRIDF ANDKFIGTSE YWYTANDVFK LGGEYNFDKF 360
361 SKSLREFCNS NWTQILANSD KGVYNSIPEN FLKDACFKGN WVLNILHEGF DMPRIDVDAE 420
421 NVNDRPLFQS VEKVEERELS WTLGRILLYA SGSILAGNDD FMVGIAPSER RTKLTGKKFI 480
481 PGKLLESDQL RKQSSSLSNK GFLMWFAIIC CIFYLIFHRS HIIRRRFSGL YNITKDFKTG 540
541 IRRRLKFLRR SDPFSRLEEG ELGTDVDGFK DVYRMKSSSM FDLGKSSATM QREHEPQRTA 600
601 SQSANLAPSN LRPAFSMADF SKFKDSRLYD |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Qiu et al. (2008) |
|
GDA1 YND1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
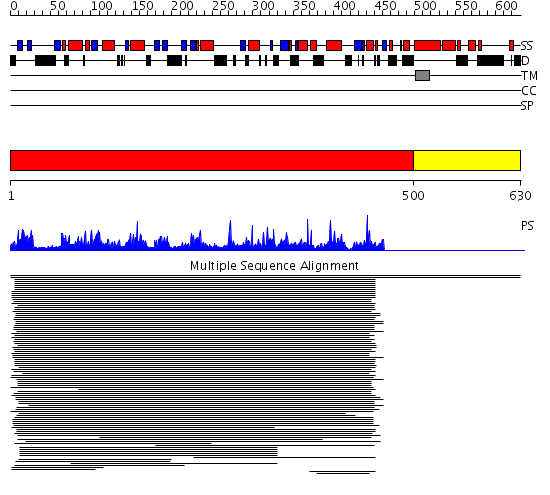
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..499] | 287.420216 | GDA1/CD39 (nucleoside phosphatase) family No confident structure predictions are available. | |
| 2 | View Details | [500..630] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [384..450] | 4.120996 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [451..630] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|