













| General Information: |
|
| Name(s) found: |
AFG1 /
YEL052W
[SGD]
|
| Description(s) found:
Found 23 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 509 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrion
[IDA |
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
ATPase activity
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MIALKPNAVR TFRQVQHCSF RICRYQSTKS NKCLTPLQEY DRLVKLGKLR DDTYQRGIIS 60
61 SLGDLYDSLV KYVPPVVKTP NAVDQVGGWL NGLKSVFSRG KPKNIGAYVD VSKIGNSIPR 120
121 GVYLYGDVGC GKTMLMDLFY TTIPNHLTKK RIHFHQFMQY VHKRSHEIVR EQNLKELGDA 180
181 KGKEIDTVPF LAAEIANNSH VLCFDEFQVT DVADAMILRR LMTALLSDDY GVVLFATSNR 240
241 HPDELYINGV QRQSFIPCIE LIKHRTKVIF LNSPTDYRKI PRPVSSVYYF PSDTSIKYAS 300
301 KECKTRRETH IKEWYNYFAQ ASHTDDSTDS HTVHKTFYDY PLTIWGREFK VPKCTPPRVA 360
361 QFTFKQLCGE PLAAGDYLTL AKNFEAFIVT DIPYLSIYVR DEVRRFITFL DAVYDSGGKL 420
421 ATTGAADFSS LFVEPEQILN DFELRPTTKE PDSVDTGMVD EMVEKHGFSK EIAKKSQMFA 480
481 LDEERFAFAR ALSRLSQMSS TDWVTKPTY |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
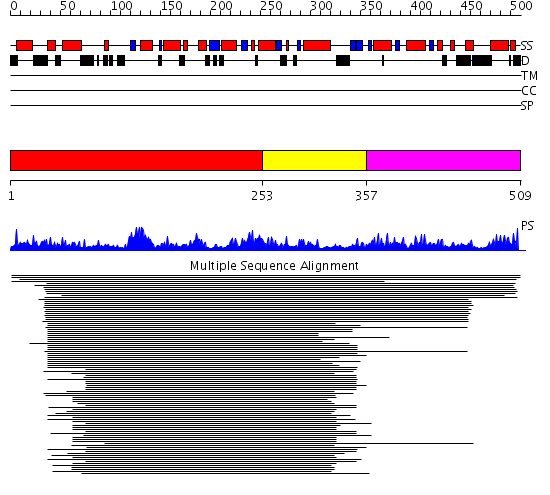
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..252] | 56.30103 | Membrane fusion atpase p97 N-terminal domain , P97-Nn; Membrane fusion atpase p97, D1 domain; Membrane fusion atpase p97 domain 2, P97-Nc | |
| 2 | View Details | [253..356] | 56.30103 | Membrane fusion atpase p97 N-terminal domain , P97-Nn; Membrane fusion atpase p97, D1 domain; Membrane fusion atpase p97 domain 2, P97-Nc | |
| 3 | View Details | [357..509] | 1.097982 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)