













| General Information: |
|
| Name(s) found: |
STN1 /
YDR082W
[SGD]
|
| Description(s) found:
Found 34 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 494 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nuclear telomere cap complex
[TAS |
| Biological Process: |
telomere capping
[TAS |
| Molecular Function: |
protein binding
[IDA telomeric DNA binding [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDKYGHIAHQ EGDVCYYIPR LFKYNSYYSG TEDVRIFVGD LKYRMRVSLQ ICEKYYDRRL 60
61 SMLFWKNHPL QQIHLIGCII GLQFKWIGKQ EYIFFQLDDC TSDSSLVGYT SDMRFLTCKV 120
121 KKDSILSWGL NITDLIGLTL HVYGQASLNY QELQVEYLRL CYSLTEEIDH WKITMNMREQ 180
181 LDTPWSLSDF VIGELFTQEQ EWTPETSQIE VVNPDFVGIG YKTPESKRNE TTFIEQLQEE 240
241 RLKDELEIIS PYNSTDTSNS VHSLSFRFVS SLKDFPETHF LNSGDQIDNG NDEQLKKLEY 300
301 QSANLPVMIP NRTSAKSNLM LILLGLQMKE ISNSDLYKLK EVRSVVTSLA SFLFQQQNVG 360
361 VMKSFDSLEK EAFRDLVNRL VSQGLIGLKD KTSETFDLLP LKNLFEYAEK RISVLMKLQC 420
421 YTGTVQLSHV QEKLHLPYIT TNGIVDVFKE CLKRTKKQYP EVLKNWWIDL DPKNGMEDQN 480
481 SGILLHLEYA AAYS |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Qiu et al. (2008) |
|
CDC13 RAP1 STN1 TEN1 |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Uetz P, et al. (2000) |
New Feature: Upload Your Own Microscopy Data
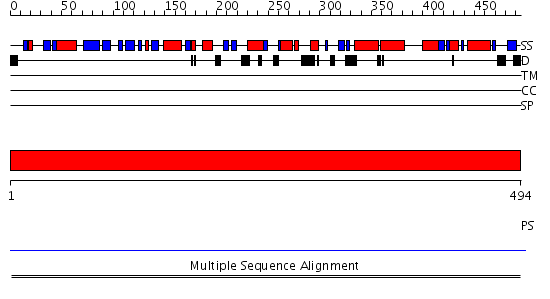
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..494] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [311..494] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.76 |
Source: Reynolds et al. (2008)