













| General Information: |
|
| Name(s) found: |
LEU2 /
YCL018W
[SGD]
|
| Description(s) found:
Found 28 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 364 amino acids |
Gene Ontology: |
|
| Cellular Component: |
soluble fraction
[IDA |
| Biological Process: |
leucine biosynthetic process
[IMP |
| Molecular Function: |
3-isopropylmalate dehydrogenase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSAPKKIVVL PGDHVGQEIT AEAIKVLKAI SDVRSNVKFD FENHLIGGAA IDATGVPLPD 60
61 EALEASKKAD AVLLGAVGGP KWGTGSVRPE QGLLKIRKEL QLYANLRPCN FASDSLLDLS 120
121 PIKPQFAKGT DFVVVRELVG GIYFGKRKED DGDGVAWDSE QYTVPEVQRI TRMAAFMALQ 180
181 HEPPLPIWSL DKANVLASSR LWRKTVEETI KNEFPTLKVQ HQLIDSAAMI LVKNPTHLNG 240
241 IIITSNMFGD IISDEASVIP GSLGLLPSAS LASLPDKNTA FGLYEPCHGS APDLPKNKVN 300
301 PIATILSAAM MLKLSLNLPE EGKAIEDAVK KVLDAGIRTG DLGGSNSTTE VGDAVAEEVK 360
361 KILA |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Qiu et al. (2008) |

|
LEU1 LEU2 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Cheeseman IM, et al. (2002) | |
| View Run | No Comments | Cheeseman IM, et al. (2002) | |
| View Run | No Comments | Cheeseman IM, et al. (2002) | |
| View Run | No Comments | Cheeseman IM, et al. (2002) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
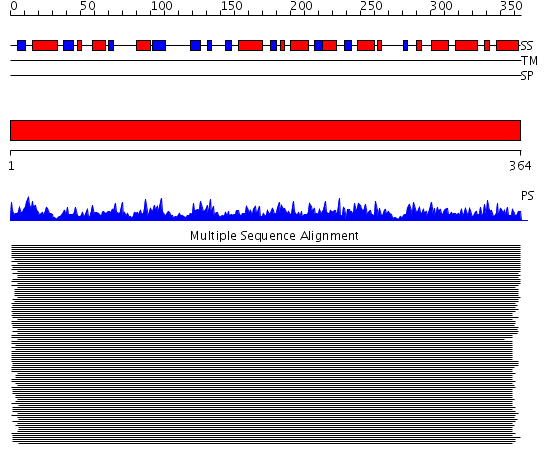
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..364] | 954.54902 | 3-isopropylmalate dehydrogenase, IPMDH |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)