













| General Information: |
|
| Name(s) found: |
QRI1 /
YDL103C
[SGD]
|
| Description(s) found:
Found 29 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 477 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [IDA |
| Biological Process: |
UDP-N-acetylglucosamine biosynthetic process
[IDA |
| Molecular Function: |
UDP-N-acetylglucosamine diphosphorylase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTDTKQLFIE AGQSQLFHNW ESLSRKDQEE LLSNLEQISS KRSPAKLLED CQNAIKFSLA 60
61 NSSKDTGVEI SPLPPTSYES LIGNSKKENE YWRLGLEAIG KGEVAVILMA GGQGTRLGSS 120
121 QPKGCYDIGL PSKKSLFQIQ AEKLIRLQDM VKDKKVEIPW YIMTSGPTRA ATEAYFQEHN 180
181 YFGLNKEQIT FFNQGTLPAF DLTGKHFLMK DPVNLSQSPD GNGGLYRAIK ENKLNEDFDR 240
241 RGIKHVYMYC VDNVLSKIAD PVFIGFAIKH GFELATKAVR KRDAHESVGL IATKNEKPCV 300
301 IEYSEISNEL AEAKDKDGLL KLRAGNIVNH YYLVDLLKRD LDQWCENMPY HIAKKKIPAY 360
361 DSVTGKYTKP TEPNGIKLEQ FIFDVFDTVP LNKFGCLEVD RCKEFSPLKN GPGSKNDNPE 420
421 TSRLAYLKLG TSWLEDAGAI VKDGVLVEVS SKLSYAGENL SQFKGKVFDR SGIVLEK |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |
|
GUS1 QRI1 SLX4 UBA4 YOR097C |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
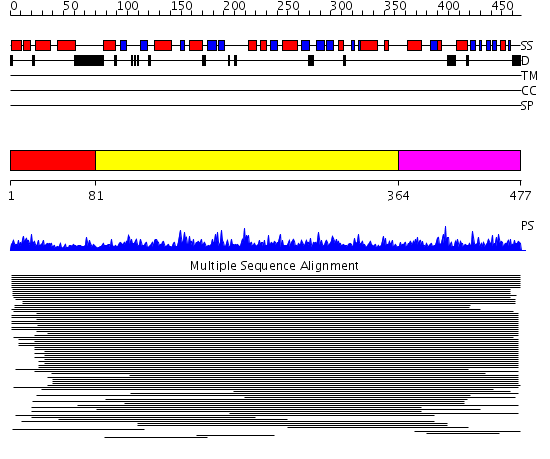
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..80] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [81..363] | 10.71 | N-acetylglucosamine 1-phosphate uridyltransferase GlmU, C-terminal domain; N-acetylglucosamine 1-phosphate uridyltransferase GlmU, N-terminal domain | |
| 3 | View Details | [364..477] | 2.044983 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)