













| General Information: |
|
| Name(s) found: |
par-1-PH /
FBpp0085653
[FlyBase]
|
| Description(s) found:
Found 36 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 993 amino acids |
Gene Ontology: |
|
| Cellular Component: |
germline ring canal
[IDA]
cell cortex [IDA] fusome [IDA] basolateral plasma membrane [IDA] |
| Biological Process: |
regulation of cell shape
[IMP]
germarium-derived oocyte fate determination [IMP][TAS] axis specification [IMP] border follicle cell delamination [IMP] actin filament organization [IMP] gamete generation [IMP] border follicle cell migration [IMP] oocyte differentiation [TAS] microtubule cytoskeleton organization [IMP] protein amino acid phosphorylation [ISS][NAS] oocyte anterior/posterior axis specification [TAS] anterior/posterior axis specification [TAS] maintenance of protein location [TAS] regulation of pole plasm oskar mRNA localization [IMP] pole plasm protein localization [IMP] oocyte nucleus localization involved in oocyte dorsal/ventral axis specification [IMP] ovarian follicle cell development [IMP] pole plasm oskar mRNA localization [TAS] regulation of Wnt receptor signaling pathway [TAS] antimicrobial humoral response [IMP] oocyte microtubule cytoskeleton organization [IMP][TAS] |
| Molecular Function: |
tau-protein kinase activity
[IDA]
ATP binding [IEA] protein kinase activity [IDA] protein serine/threonine kinase activity [ISS][NAS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGQTSSHRQM HHSSNNNNKE EPPQPASQFG NRRTSTRKST SSSKHYPTLV QPLRKDELSL 60
61 LRDSLLRNGS IMRKNPERRP VTISELKPVN TGQGTSGSVL PITAEDAAGG SSSSITQLMA 120
121 FQLPVDRSAK TDDLAAESSA SAAAAVAPKE GRNEFYIVGD GYHRTDHCYY RSPNGDFHKL 180
181 PSDSFHKMSE GCFVRMADGV FRRLEVPTKA QISRRSSRAE EIGDATSQGN GSAATNVKSQ 240
241 LMRFLKRSKS HTPATIAKLQ REKEEARAER KQRLSNALRQ KNLAATNAAS AAAANSSYAS 300
301 GGGGGTVSSR NNKIVVTMIE NGGLPIVATS KAERPKAKES ASSSDKARNS RGSPNMQMRS 360
361 SAPMRWRATE EHIGKYKLIK TIGKGNFAKV KLAKHLPTGK EVAIKIIDKT QLNPGSLQKL 420
421 FREVRIMKML DHPNIVKLFQ VIETEKTLYL IMEYASGGEV FDYLVLHGRM KEKEARVKFR 480
481 QIVSAVQYCH QKRIIHRDLK AENLLLDSEL NIKIADFGFS NEFTPGSKLD TFCGSPPYAA 540
541 PELFQGKKYD GPEVDVWSLG VILYTLVSGS LPFDGSTLRE LRERVLRGKY RIPFYMSTDC 600
601 ENLLRKFLVL NPAKRASLET IMGDKWMNMG FEEDELKPYI EPKADLADPK RIEALVAMGY 660
661 NRSEIEASLS QVRYDDVFAT YLLLGRKSTD PESDGSRSGS SLSLRNISGN DAGANAGSAS 720
721 VQSPTHRGVH RSISASSTKP SRRASSGAET LRVGPTNAAA TVAAATGAVG AVNPSNNYNA 780
781 AGSAADRASV GSNFKRQNTI DSATIKENTA RLAAQNQRPA SATQKMLTTA DTTLNSPAKP 840
841 RTATKYDPTN GNRTVSGTSG IIPRRSTTLY EKTSSTEKTN VIPAETNSGS AAPAGKGHTK 900
901 SASVSSPRPS TDGCVSVSND PLGTRMASAV KSSRHFPRNV PSRSTFHSGQ TRARNNTALE 960
961 YSGTSGASGD SSHPGRMSFF SKLSSRFSKR PNQ |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
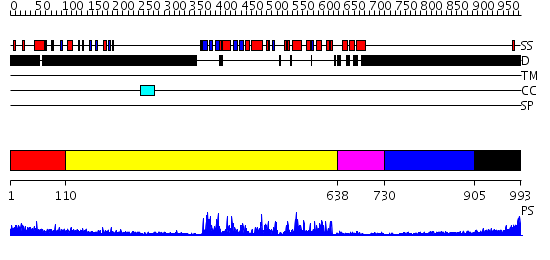
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..109] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [110..637] | 59.221849 | No description for 2h8hA was found. | |
| 3 | View Details | [638..729] | 100.0 | No description for 2qnjA was found. | |
| 4 | View Details | [730..904] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [905..993] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)