













| General Information: |
|
| Name(s) found: |
C15-PA /
FBpp0083510
[FlyBase]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 339 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[ISS]
protein complex [IPI] |
| Biological Process: |
imaginal disc-derived leg morphogenesis
[IMP]
negative regulation of transcription [IMP] regulation of transcription, DNA-dependent [IEA][ISS] leg disc development [IMP] positive regulation of transcription [IMP] antennal morphogenesis [IMP] bristle development [IMP] regulation of transcription [IEA] elongation of arista core [IMP] positive regulation of Notch signaling pathway [IEP] mesodermal cell fate specification [NAS] |
| Molecular Function: |
transcription factor activity
[ISS]
sequence-specific DNA binding [IPI] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSHEEDDHE HENEHGEEVE EQEIHVDVDS DSRMSCGSGS DVDMDGGSCY DESETPLSES 60
61 LQSEQTRSSS SENLPFSISR LLSKPFETSH HHHNNNNHLL SSSPGSSSNN NNSGEKGEEK 120
121 ELLQQEDHDL AYKLATSIAN STYGSAAALY SYPHLYPSAA GGHVLRVPPQ RTPLTWALPP 180
181 LHHAALAHQA VKDRLAAAFP IARRIGHPYQ NRTPPKRKKP RTSFTRIQVA ELEKRFHKQK 240
241 YLASAERAAL ARGLKMTDAQ VKTWFQNRRT KWRRQTAEER EAERQAANRL MLSLQAEAIS 300
301 KGFAPPSAPL GSQGGVNGAP LAALHGLQPW AEASHAAGC |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
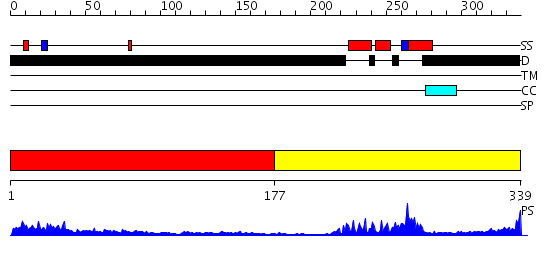
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..176] | 5.897996 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [177..339] | 27.09691 | DETERMINATION OF THE THREE-DIMENSIONAL STRUCTURE OF THE ANTENNAPEDIA HOMEODOMAIN FROM DROSOPHILA IN SOLUTION BY 1H NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)