













| General Information: |
|
| Name(s) found: |
sol-PB /
FBpp0088620
[FlyBase]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1594 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[NAS]
intracellular [IEA] |
| Biological Process: |
proteolysis
[IEA]
nervous system development [IMP] visual behavior [NAS] |
| Molecular Function: |
zinc ion binding
[IEA]
calcium-dependent cysteine-type endopeptidase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGTISSVLQW SCTKCNTINP TESLKCFNCG TVRKVFPQQQ QQQHRSSSIT ASWTADDALE 60
61 QEQAEKGQER DKEKGRAAVA RSEYKHVYKS LLRGCLKRPQ RNSQNLPANC VDCEDTRKYI 120
121 KSSIELYRHF SNPALNRRWV CHACGTDNSS VTWHCLICDT VSYLAPIYKD AIAADRGQDL 180
181 AGSLGNRGEL LAADHSHPHH HHHYLHQELE EQHQHQLHSQ HLHKRHLKGR SASGSGSGPG 240
241 SGSGLRRTQS LSTAIDKSAS GRSCHICYAN NQSKDIFNLP QIKPAPQLTG IPPVAACSNS 300
301 RFAIANDTFC RRKQNNNNKN QNHKVVRESG AKRKYNFTIT TLSRSAAKDA GHGQMKPLRQ 360
361 VVNLNLNLQQ EPQQKSPANP QQLQRKTQRE PAAVSMNPTQ FTIPRNGVFI AVNEWSEPMA 420
421 SSSSVSSSSN HHHHHHSNSN SNSSGNSNII NNNSSSSSGS NKLYENECVA LAQQQLRAAA 480
481 AQAAQAAATA VAIASSPSAK AMAEPAPTAT MPIYAQVNKQ HKLKKKQQIA SESQTNNNTG 540
541 SGEIADAVSE SLTAGLGTST DGSGEASESE SQVEEHSIYA KVWKGPRKAT ESKIMHDPGS 600
601 SSRLSGAASA AAGTASAGAI AAAVGAAAAS RHDNKTQLGN GSRSKMWICI KCSYAYNRLW 660
661 LQTCEMCEAK AEQQQQQLQL QQQQQQQQQH HHHHLQQQQA EAPRDEPWTC KKCTLVNYST 720
721 AMACVVCGGS KLKSISSIED MTLRKGEFWT CSHCTLKNSL HSPVCSACKS HRQPQLSMAM 780
781 EAVRERPDGQ SYEEQDAAAV GGGGGSAHQS GANEVKAPTA LNLPLTSVAL PMPMLQLPTS 840
841 TAAGLRGSRS PSPRMQLLPS LQQQRNSSSS GAIPKRHSTG GSIVPRNISI AGLANYNLQQ 900
901 GQGVGSASVV SASGAGSGAG AVGASTSTKK WQCPACTYDN CAASVVCDIC SSPRGLASAV 960
961 LGEALGRKSV RVALTPADIR QESKLMENLR QLEETEALTK WQNIIQYCRD NSELFVDDSF1020
1021 PPAPKSLYYN PASGAGEGNP VVQWRRPHEI NCDGGAYPPW AVFRTPLPSD ICQGVLGNCW1080
1081 LLSALAVLAE REDLVKEVLV TKEICGQGAY QVRLCKDGKW TTVLVDDLLP CDKRGHLVYS1140
1141 QAKRKQLWVP LIEKAVAKIH GCYEALVSGR AIEGLATLTG APCESIPLQA SSLPMPSEDE1200
1201 LDKDLIWAQL LSSRCVRFLM GASCGGGNMK VDEEEYQQKG LRPRHAYSVL DVKDIQGHRL1260
1261 LKLRNPWGHY SWRGDWSDDS SLWTDDLRDA LMPHGASEGV FWISFEDVLN YFDCIDICKV1320
1321 RSGWNEVRLQ GTLQPLCSIS CVLLTVLEPT EAEFTLFQEG QRNSEKSQRS QLDLCVVIFR1380
1381 TRSPAAPEIG RLVEHSKRQV RGFVGCHKML ERDIYLLVCL AFNHWHTGIE DPHQYPQCIL1440
1441 AIHSSKRLLV EQISPSPHLL ADAIISLTLT KGQRHEGREG MTAYYLTKGW AGLVVMVENR1500
1501 HENKWIHVKC DCQESYNVVS TRGELKTVDS VPPLQRQVII VLTQLEGSGG FSIAHRLTHR1560
1561 LANSRGLHDW GPPGATHCPP IENVHGLHAP RLIT |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
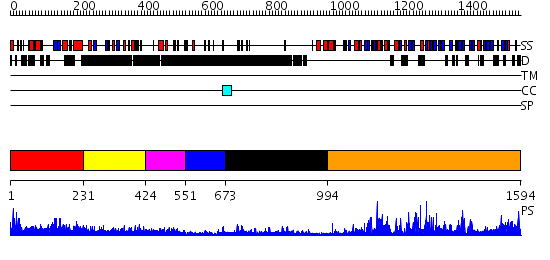
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..230] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [231..423] | 3.281993 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [424..550] | 3.293989 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [551..672] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [673..993] | 3.69897 | Receptor protein-tyrosine kinase Erbb-3 extracellular domain; Receptor protein-tyrosine kinase Erbb-3 Cys-rich domains | |
| 6 | View Details | [994..1594] | 1000.0 | Crystal Structure of a mu-like calpain |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||
| 1 | No functions predicted. | |||||||||||||||
| 2 |
|
|||||||||||||||
| 3 | No functions predicted. | |||||||||||||||
| 4 | No functions predicted. | |||||||||||||||
| 5 |
|
|||||||||||||||
| 6 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.84 |
Source: Reynolds et al. (2008)