













| General Information: |
|
| Name(s) found: |
CG9517-PA /
FBpp0073799
[FlyBase]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Drosophila melanogaster |
| Length: | 865 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
alcohol metabolic process
[IEA]
|
| Molecular Function: |
FAD binding
[IEA]
glucose dehydrogenase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGQEISQQQN QQHQQHQQHQ QQQLPHHKRQ PQRKSFSGIS TTNQPIYTPT SHLSTSSSSI 60
61 NTNDYSHNNT SNNNNNNQNY RLRHKYNLSK QTYERSKIAA SSASTSQSDT DSHYAKLNLQ 120
121 RSSSQSSDSG IYNSSCHCSS ISSISAVSSS ASSSRSSSSS SRGCPSSVVS SKSNRSLDKQ 180
181 KHYLSHHLYI KSYLDILEED ERARAHFKSA RPGGIRNYTP KILASGESSL SGNSSTNTAL 240
241 SSSAPAHCYG PGKRTTVGGT SGSGGAAGAT GTGAQGTDPV DPENKVQEPT VIRRQYDFVV 300
301 IGGGSAGAVV ANRLSEVRNW TVLLLEAGGD ETEISDVPAL AGYLQLTELD WKYQTTPSST 360
361 RQYCQAMKGD RCFWPRGKVL GGSSVLNAMV YVRGSKNDYN HWASLGNPGW DYDSMLKYFL 420
421 KSEDVRNPYL AKTPYHETGG YLTVQEAPWR TPLSIAFLQA GIEMGYENRD INGAQQTGFM 480
481 LTQSTIRRGA RCSTGKAFIR PVRQRKNFDV LLHAEATRIL FDKQKRAIGV EYMRGGRKNV 540
541 VFVRREVIAS AGALNTPKLL MLSGVGPAEH LQEHNIPVIS DLPVGNNMQD HVGLGGLTFV 600
601 VDAPLTVTRN RFQTIPVSME YILRERGPMT FSGVEGVAFL NTKYQDPSVD WPDVQFHFCP 660
661 SSINSDGGEQ IRKILNLRDG FYNTVYKPLQ HSETWSILPL LLRPKSTGWV RLNSRNPQHQ 720
721 PKIIPNYFAH QEDIDVLVEG IKLAINVSNT QAFQRFGSRL HNIPLPGCRH LPFQSNEYWA 780
781 CCIKEFTFTI YHPAGTCRMG PSWDVTAVVD PRLRVYGVSG VRVVDASIMP TIVNGNPNAP 840
841 VIAIGEKASD LIKEDWGVRR AHTSA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
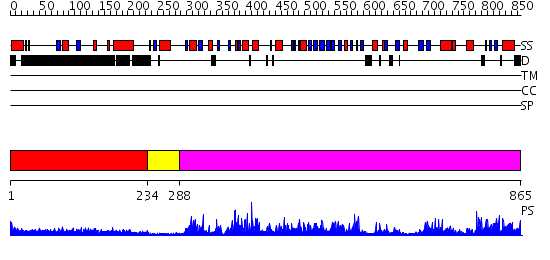
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..233] | 4.045757 | Botulinum neurotoxin | |
| 2 | View Details | [234..287] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [288..865] | 105.0 | Glucose oxidase |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)