













| General Information: |
|
| Name(s) found: |
dwg-PA /
FBpp0070465
[FlyBase]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 592 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[ISS]
nuclear chromatin [IDA] |
| Biological Process: | NONE FOUND |
| Molecular Function: |
transcription factor activity
[ISS]
zinc ion binding [IEA] protein binding [IPI] chromatin insulator sequence binding [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MMNSKIAEVV VLNCRTCTRA CKLHKPLQEE IDLGSEGSTT LASMLNYCTG LSFEPQDGAA 60
61 MPQHICLHCL QLLEQAFNFK RMVIDSDELL RLGLDEARCS SFHESQTHSP NQSQQHDQQQ 120
121 QAFDSTEEYV MIEMLNEEQE NVANLDEELA EEDRRIFAMT VDEEEEFLTE EVDQDDELSE 180
181 EEHQLAQSGD QQEEHITMES VHEFQPAEVE YVTIKNEFET IVTEEDEFEV MNDSGAHDAI 240
241 LDCQMIVIPA EGAIDEVIGE ETLELEGDGR EEHLLPEAED VCEDEDFLEE SLDSAPPTAG 300
301 EALPYVCTVC QKAFRQQCRL NQHMRSHVDE KQYECEECGK RLKHLRNYKE HMLTHTNVKP 360
361 HQCSICGRFY RTTSSLAVHK RTHAEKKPYN CDQCGRGYAA FDHLRRHKLT HTGERPYACD 420
421 LCDKAYYDSS SLRQHKISHT GKKAFTCEIC GVGLSQKSGY KKHMMVHSGV KAHKCDVCGH 480
481 AFTFTSNLNA HVRLHSGEKP FKCEVCVKAF PTKKRLASHM RVHNKESPVT ATVAVQSINP 540
541 PSRQAGAEGA TGGGATGGSP TGGGATGGSA TNLHITEVHE QPVSTSKVVM VL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
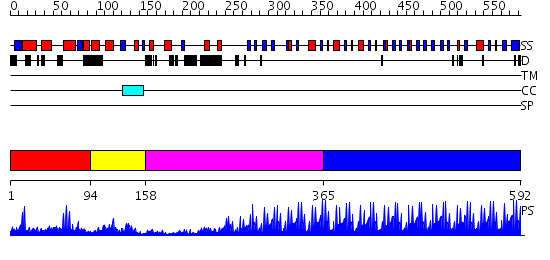
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..93] | 7.0 | Crystal structure of the zinc finger associated domain of the Drosophila transcription factor Grauzone | |
| 2 | View Details | [94..157] | 10.522879 | Solution structure of the Kruppel-associated box (KRAB) domain | |
| 3 | View Details | [158..364] | 7.6 | No description for 2i13A was found. | |
| 4 | View Details | [365..592] | 66.69897 | No description for 2i13A was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)