













| General Information: |
|
| Name(s) found: |
NIFJ_ECOLI
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Escherichia coli |
| Length: | 1174 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MITIDGNGAV ASVAFRTSEV IAIYPITPSS TMAEQADAWA GNGLKNVWGD TPRVVEMQSE 60
61 AGAIATVHGA LQTGALSTSF TSSQGLLLMI PTLYKLAGEL TPFVLHVAAR TVATHALSIF 120
121 GDHSDVMAVR QTGCAMLCAA NVQEAQDFAL ISQIATLKSR VPFIHFFDGF RTSHEINKIV 180
181 PLADDTILDL MPQVEIDAHR ARALNPEHPV IRGTSANPDT YFQSREATNP WYNAVYDHVE 240
241 QAMNDFSAAT GRQYQPFEYY GHPQAERVII LMGSAIGTCE EVVDELLTRG EKVGVLKVRL 300
301 YRPFSAKHLL QALPGSVRSV AVLDRTKEPG AQAEPLYLDV MTALAEAFNN GERETLPRVI 360
361 GGRYGLSSKE FGPDCVLAVF AELNAAKPKA RFTVGIYDDV TNLSLPLPEN TLPNSAKLEA 420
421 LFYGLGSDGS VSATKNNIKI IGNSTPWYAQ GYFVYDSKKA GGLTVSHLRV SEQPIRSAYL 480
481 ISQADFVGCH QLQFIDKYQM AERLKPGGIF LLNTPYSADE VWSRLPQEVQ AVLNQKKARF 540
541 YVINAAKIAR ECGLAARINT VMQMAFFHLT QILPGDSALA ELQGAIAKSY SSKGQDLVER 600
601 NWQALALARE SVEEVPLQPV NPHSANRPPV VSDAAPDFVK TVTAAMLAGL GDALPVSALP 660
661 PDGTWPMGTT RWEKRNIAEE IPIWKEELCT QCNHCVAACP HSAIRAKVVP PEAMENAPAS 720
721 LHSLDVKSRD MRGQKYVLQV APEDCTGCNL CVEVCPAKDR QNPEIKAINM MSRLEHVEEE 780
781 KINYDFFLNL PEIDRSKLER IDIRTSQLIT PLFEYSGACS GCGETPYIKL LTQLYGDRML 840
841 IANATGCSSI YGGNLPSTPY TTDANGRGPA WANSLFEDNA EFGLGFRLTV DQHRVRVLRL 900
901 LDQFADKIPA ELLTALKSDA TPEVRREQVA ALRQQLNDVA EAHELLRDAD ALVEKSIWLI 960
961 GGDGWAYDIG FGGLDHVLSL TENVNILVLD TQCYSNTGGQ ASKATPLGAV TKFGEHGKRK1020
1021 ARKDLGVSMM MYGHVYVAQI SLGAQLNQTV KAIQEAEAYP GPSLIIAYSP CEEHGYDLAL1080
1081 SHDQMRQLTA TGFWPLYRFD PRRADEGKLP LALDSRPPSE APEETLLHEQ RFRRLNSQQP1140
1141 EVAEQLWKDA AADLQKRYDF LAQMAGKAEK SNTD |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
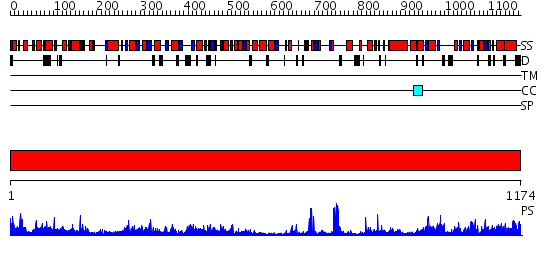
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..1174] | 1000.0 | Pyruvate-ferredoxin oxidoreductase, PFOR, domain I; Pyruvate-ferredoxin oxidoreductase, PFOR, domains VI; Pyruvate-ferredoxin oxidoreductase, PFOR, domain II; Pyruvate-ferredoxin oxidoreductase, PFOR, domain III; Pyruvate-ferredoxin oxidoreductase, PFOR, domain V |
Functions predicted (by domain):
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.91 |
Source: Reynolds et al. (2008)