













| General Information: |
|
| Name(s) found: |
MUTL_ECOLI
[Swiss-Prot]
|
| Description(s) found:
Found 13 descriptions. SHOW ALL |
|
| Organism: | Escherichia coli |
| Length: | 615 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPIQVLPPQL ANQIAAGEVV ERPASVVKEL VENSLDAGAT RIDIDIERGG AKLIRIRDNG 60
61 CGIKKDELAL ALARHATSKI ASLDDLEAII SLGFRGEALA SISSVSRLTL TSRTAEQQEA 120
121 WQAYAEGRDM NVTVKPAAHP VGTTLEVLDL FYNTPARRKF LRTEKTEFNH IDEIIRRIAL 180
181 ARFDVTINLS HNGKIVRQYR AVPEGGQKER RLGAICGTAF LEQALAIEWQ HGDLTLRGWV 240
241 ADPNHTTPAL AEIQYCYVNG RMMRDRLINH AIRQACEDKL GADQQPAFVL YLEIDPHQVD 300
301 VNVHPAKHEV RFHQSRLVHD FIYQGVLSVL QQQLETPLPL DDEPQPAPRS IPENRVAAGR 360
361 NHFAEPAARE PVAPRYTPAP ASGSRPAAPW PNAQPGYQKQ QGEVYRQLLQ TPAPMQKLKA 420
421 PEPQEPALAA NSQSFGRVLT IVHSDCALLE RDGNISLLSL PVAERWLRQA QLTPGEAPVC 480
481 AQPLLIPLRL KVSAEEKSAL EKAQSALAEL GIDFQSDAQH VTIRAVPLPL RQQNLQILIP 540
541 ELIGYLAKQS VFEPGNIAQW IARNLMSEHA QWSMAQAITL LADVERLCPQ LVKTPPGGLL 600
601 QSVDLHPAIK ALKDE |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
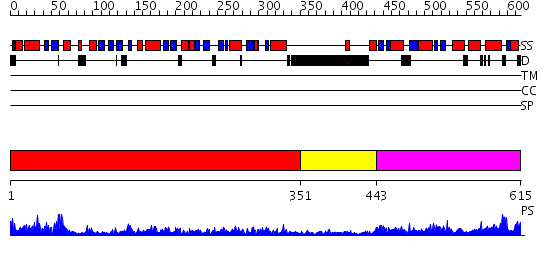
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..350] | 87.69897 | DNA mismatch repair protein MutL | |
| 2 | View Details | [351..442] | 18.221849 | No description for 2cgeA was found. | |
| 3 | View Details | [443..615] | 28.522879 | Crystal structure of the MutL C-terminal domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)