













| General Information: |
|
| Name(s) found: |
ISPG_ECOLI
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Escherichia coli |
| Length: | 372 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MHNQAPIQRR KSTRIYVGNV PIGDGAPIAV QSMTNTRTTD VEATVNQIKA LERVGADIVR 60
61 VSVPTMDAAE AFKLIKQQVN VPLVADIHFD YRIALKVAEY GVDCLRINPG NIGNEERIRM 120
121 VVDCARDKNI PIRIGVNAGS LEKDLQEKYG EPTPQALLES AMRHVDHLDR LNFDQFKVSV 180
181 KASDVFLAVE SYRLLAKQID QPLHLGITEA GGARSGAVKS AIGLGLLLSE GIGDTLRVSL 240
241 AADPVEEIKV GFDILKSLRI RSRGINFIAC PTCSRQEFDV IGTVNALEQR LEDIITPMDV 300
301 SIIGCVVNGP GEALVSTLGV TGGNKKSGLY EDGVRKDRLD NNDMIDQLEA RIRAKASQLD 360
361 EARRIDVQQV EK |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
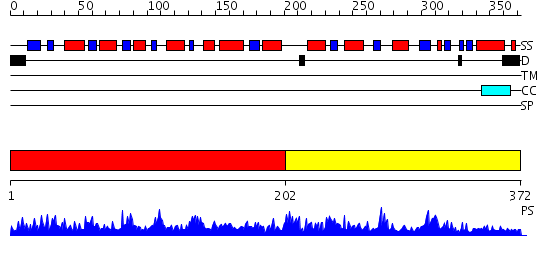
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..201] | 1.53 | Crystal structure of a cyclase subunit of imidazolglycerolphosphate synthase | |
| 2 | View Details | [202..372] | 31.0 | SULFITE REDUCTASE STRUCTURE AT 1.6 ANGSTROM RESOLUTION |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)