













| General Information: |
|
| Name(s) found: |
gi|151944013
[NCBI NR]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae YJM789 |
| Length: | 359 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSEVITITKR NGAFQNSSNL SYNNTGISDD ENDEEDIYMH DVNSASKSES DSQIVTPGEL 60
61 VTDDPIWMRG HGTYFLDNMT YSSVAGTVSR VNRLLSVIPL KGRYAPETGD HVVGRIAEVG 120
121 NKRWKVDIGG KQHAVLMLGS VNLPGGILRR KSESDELQMR SFLKEGDLLN AEVQSLFQDG 180
181 SASLHTRSLK YGKLRNGMFC QVPSSLIVRA KNHTHNLPGN ITVVLGVNGY IWLRKTSQMD 240
241 LARDTPSANN SSSIKSTGPT GAVSLNPSIT RLEEESSWQI YSDENDPSIS NNIRQAICRY 300
301 ANVIKALAFC EIGITQQRIV SAYEASMVYS NVGELIEKNV MESIGSDILT AEKMRGNGN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
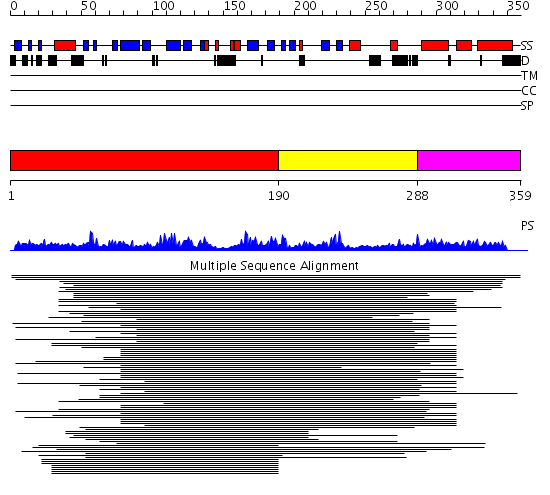
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..189] | 23.69897 | Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domain 3; S1 RNA-binding domain of polyribonucleotide phosphorylase, PNPase; Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domains 1 and 4; Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domain 6; Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domains 2 and 5 | |
| 2 | View Details | [190..287] | 2.200996 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [288..359] | N/A | Confident ab initio structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)