













| General Information: |
|
| Name(s) found: |
HSD17B1
[HGNC (HUGO)]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Homo sapiens |
| Length: | 327 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 ARTVVLITGC SSGIGLHLAV RLASDPSQSF KVYATLRDLK TQGRLWEAAR ALACPPGSLE 60
61 TLQLDVRDSK SVAAARERVT EGRVDVLVCN AGLGLLGPLE ALGEDAVASV LDVNVVGTVR 120
121 MLQAFLPDMK RRGSGRVLVT GSVGGLMGLP FNDVYCASKF ALEGLCESLA VLLLPFGVHL 180
181 SLIECGPVHT AFMEKVLGSP EEVLDRTDIH TFHRFYQYLA HSKQVFREAA QNPEEVAEVF 240
241 LTALRAPKPT LRYFTTERFL PLLRMRLDDP SGSNYVTAMH REVFGDVPAK AEAGAEAGGG 300
301 AGPGAEDEAG RSAVGDPELG DPPAAPQ |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Noble WS, et al. (2009) Manuscript in preparation. |

|
HSD17B1 gi|112491310 |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
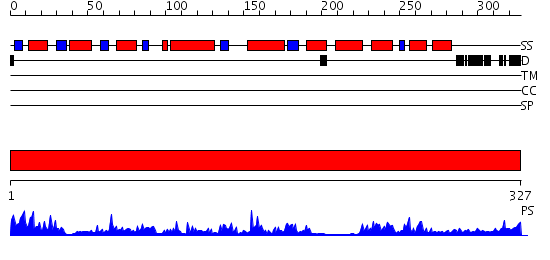
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..327] | 56.69897 | HUMAN ESTROGENIC 17BETA-HYDROXYSTEROID DEHYDROGENASE |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.70 |
Source: Reynolds et al. (2008)