













| General Information: |
|
| Name(s) found: |
CE36601
[WormBase]
|
| Description(s) found:
Found 14 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 532 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
collagen and cuticulin-based cuticle attachment to epithelium
[IMP locomotory behavior [IMP positive regulation of growth rate [IMP embryonic development ending in birth or egg hatching [IMP post-embryonic body morphogenesis [IMP gamete generation [IMP reproduction [IMP |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSDESEALLR VDHVKYRKAG DGKSPIGRLA LFTDFIEWRD NASPEVFTCK FIRINGQRVS 60
61 PPHKSKVQLQ LILKNEDQAT FVFLNPSASK EDLVKERDAV KEALQSALVK HRHRVNELAK 120
121 SVESQSKQVE LQAKQKILQE DRNLEKLYQN LVATKLITPD DFWSDYYQKE GVSEDKIGIN 180
181 GAFLANIVQQ EGTNGVRLNL NPEIIQAIYS TYPAVQKKNL ELVPHEMSEE NFWKKFFQSH 240
241 YFHREREVLP NPKDPFAECV RDDEEEMTKM TAEKIQRKRF DLEHIDDNGL RDFIHKSESQ 300
301 QKSAKTTLIK RCNYLSEKIL ATSWRSGEAA PTTSAAANQK KGFGVLDRLV AETETRLESE 360
361 DLATKEAEDE VQIVRISSSA KASQAKYSKP DMTRYKALVT DYFQNSSVED ILHRNEMLLD 420
421 QQEMETWEME EAMETDNNEV PPPENWCQSA GDLAKVRDVH SAVREICRQF WKSFPPIDKA 480
481 SEEKLNRMAG TLSKYRDDIR NSDISKENLT HCIQMIDLAV DKFEAYEMKK MK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
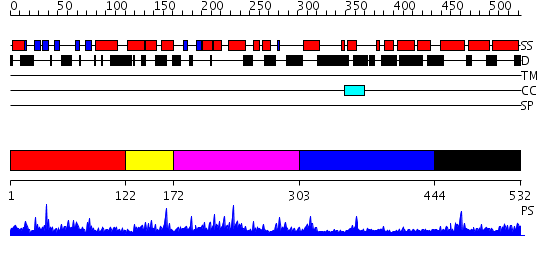
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..121] | 32.69897 | Solution structure of the N-terminal PH/PTB domain of the TFIIH P62 subunit | |
| 2 | View Details | [122..171] | 2.08 | No description for 2diiA was found. | |
| 3 | View Details | [172..302] | 17.39794 | No description for PF03909.8 was found. Confident ab initio structure predictions are available. | |
| 4 | View Details | [303..443] | 1.041989 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [444..532] | 2.039992 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)