













| General Information: |
|
| Name(s) found: |
let-413 /
CE37008
[WormBase]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 699 amino acids |
Gene Ontology: |
|
| Cellular Component: |
basolateral plasma membrane
[IDA |
| Biological Process: |
embryonic development
[IMP actin filament-based process [IMP negative regulation of multicellular organism growth [IMP hermaphrodite genitalia development [IMP locomotory behavior [IMP cell-cell junction assembly [IMP embryonic development ending in birth or egg hatching [IMP reproduction [IMP |
| Molecular Function: |
protein binding
[IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPAFFCLPMA CQRQVDSIDR SQSNLQAIPS DIFRFRKLED LNLTMNNIKE LDHRLFSLRH 60
61 LRILDVSDNE LAVLPAEIGN LTQLIELNLN RNSIAKLPDT MQNCKLLTTL NLSSNPFTRL 120
121 PETICECSSI TILSLNETSL TLLPSNIGSL TNLRVLEARD NLLRTIPLSI VELRKLEELD 180
181 LGQNELEALP AEIGKLTSLR EFYVDINSLT SLPDSISGCR MLDQLDVSEN QIIRLPENLG 240
241 RMPNLTDLNI SINEIIELPS SFGELKRLQM LKADRNSLHN LTSEIGKCQS LTELYLGQNF 300
301 LTDLPDTIGD LRQLTTLNVD CNNLSDIPDT IGNCKSLTVL SLRQNILTEL PMTIGKCENL 360
361 TVLDVASNKL PHLPFTVKVL YKLQALWLSE NQTQSILKLS ETRDDRKGIK VVTCYLLPQV 420
421 DAIDGEGRSG SAQHNTDRGA FLGGPKVHFH DQADTTFEEN KEAEIHLGNF ERHNTPHPKT 480
481 PKHKKGSIDG HMLPHEIDQP RQLSLVSNHR TSTSSFGESS NSINRDLADI RAQNGVREAT 540
541 LSPEREERMA TSLSSLSNLA AGTQNMHTIR IQKDDTGKLG LSFAGGTSND PAPNSNGDSG 600
601 LFVTKVTPGS AAYRCGLREG DKLIRANDVN MINASQDNAM EAIKKRETVE LVVLRRSPSP 660
661 VSRTSEPSLN GSSHQLNHFD AGSPDSTMFV TSSTPVYAS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
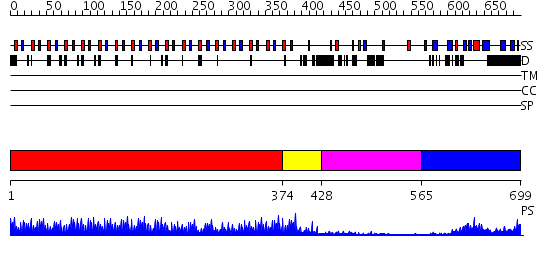
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..373] | 44.39794 | No description for 2omxA was found. | |
| 2 | View Details | [374..427] | 27.09691 | Polygalacturonase inhibiting protein PGIP | |
| 3 | View Details | [428..564] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [565..699] | 10.154902 | Erbin |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.80 |
Source: Reynolds et al. (2008)