













| General Information: |
|
| Name(s) found: |
ESM1_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 392 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
peroxisome [IDA] chloroplast [IDA] cytosolic ribosome [IDA] membrane [IDA] apoplast [IDA] chloroplast envelope [IDA] vacuole [IDA] |
| Biological Process: |
response to cold
[IEP]
glucosinolate catabolic process [IDA][IMP] response to insect [IMP] defense response to bacterium [IEP] |
| Molecular Function: |
hydrolase activity, acting on ester bonds
[IEA]
carboxylesterase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MADNLNLVSV LGVLLVLTIF HNPIIVYAGE GVPNVALFTF GDSYYDAGNK VFLSQRKDLP 60
61 QTYWPYGKSR DYPNGKFSDG HIVPDFIADF ISIPNGVLPP VLKPGVDISR GVSFAVADAS 120
121 ILGAPVESMT LNQQVVKFKN MKSNWNDSYI EKSLFMIYIG TEDYLNFTKA NPNADASAQQ 180
181 AFVTNVINRL KNDIKLLYSL GASKFVVQLL APLGCLPIVR QEYKTGNECY ELLNDLAKQH 240
241 NGKIGPMLNE FAKISTSPYG FQFTVFDFYN AVLRRIATGR SLNYRFFVTN TSCCGVGTHN 300
301 AYGCGKGNVH SKLCEYQRSY FFFDGRHNTE KAQEEMAHLL YGADPDVVQP MTVRELIVYP 360
361 TGETMREYWE PNNLAIRRRP SRDFYLGLAA YY |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
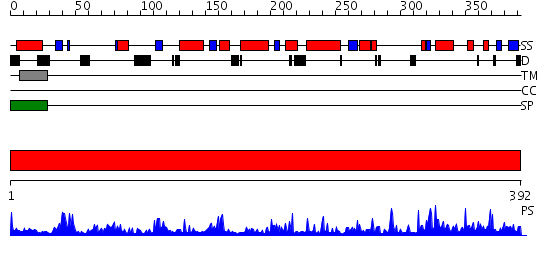
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..392] | 2.18 | THE MOLECULAR MECHANISM OF ENANTIORECOGNITION BY ESTERASES |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||
| 1 |
|
| Source: Reynolds et al. 2008. Manuscript submitted |

|