













| General Information: |
|
| Name(s) found: |
gi|9755836
[NCBI NR]
gi|63003814 [NCBI NR] gi|15237311 [NCBI NR] gi|11358175 [NCBI NR] gi|110738266 [NCBI NR] |
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 519 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
|
| Biological Process: |
nuclear-transcribed mRNA catabolic process, nonsense-mediated decay
[IMP]
negative regulation of flower development [IMP] |
| Molecular Function: |
RNA binding
[ISS]
mRNA binding [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSDSDNLQLP PSSTGATVAA TDVGWYILGE NQQNLGPYTF SELCNHFRNG YLLETTLVWA 60
61 DGRSEWQPLS AIPDLMSRIS GAEIAYPAVG SSGLINGSNA GTRQEKQDYT ASASTEDEFE 120
121 KWQREIKDAE AEAERLKNGS VSGTELVEDD HERASSPPEG EDEFTDDDGT KYKWDRARRV 180
181 WVPQDDPPLG SVDPYGLEEM TFAKEDEVFP TINILDTSVD KKDASKDDVA GKKEEDGSDE 240
241 TAEINSNGKR KLPEPETEKK EPNKPPDSWF ELKVNPHIYV NGLPDDVTIE EVAEVFSKCG 300
301 IIKEDDTGKP RIKLYSDKAT GKLKGDALIS YMKEPSVDLA IKILDGAPLR PADKLLMSVS 360
361 RAKFEQKGER FITKQTDNKK KKKLKKVEQK LLGWGGTDDS KVSIPATVVL RYMFSPAELM 420
421 ADEDLVAELE EDVKEESLKH GPFDSVKVCE HHPQGVVLVR FKDRRDAQKC IEAMNGRWYA 480
481 KRQIHASLDD GSVNHATVRD FDLEAERLDQ FAAELEAAE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
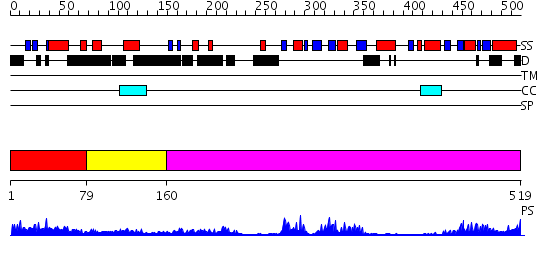
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..78] | 2.79 | Solution structure of the GYF domain of a hypothetical protein from Arabidopsis thaliana | |
| 2 | View Details | [79..159] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [160..519] | 12.0 | No description for 2ghpA was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)