













| General Information: |
|
| Name(s) found: |
PRR6_ARATH
[Swiss-Prot]
APRR6_ARATH [Swiss-Prot] |
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 663 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
two-component signal transduction system (phosphorelay)
[IEA]
regulation of transcription, DNA-dependent [IEA] regulation of transcription [IEA] |
| Molecular Function: |
transcription regulator activity
[TAS]
two-component response regulator activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAGCLVPECA DDISILLIDH DTASIASLTS MLQQFSKRVM SVDVASKALS MIEKQKKEIG 60
61 LIIANIEMPH IDSHSFLNAL LLKDIPLILI NPEIKTKEPS DLLTKRACFS LDKPISNDDI 120
121 KNMWQHVFSK KSQELKKINI TEDQENVMDK DTYQIEAFRA NLKRQRISQA SLLGRRPFID 180
181 TFSTYETFQK RKSIANVEWK TTPSYAIEIE NKRKEWKKSV GRRKSLWNSE RHMKFIAAIS 240
241 ILGEEVLFKK PIGEKEETMP KFHIGGKLDL SNHSVHGNVL NKLSMNVNFV PSTISNNPAY 300
301 NILSIDSSSI DSSSYTGLVS TGLSSENSPI LYGLPSNDGA SNTCTSQMES ERISIPQYDP 360
361 NQCHPHRSIL ETDVNQIDLD FTSILDSFDP LVDECLMKEN NRFLPNPTMN LLDTDIDKMD 420
421 WVSFIENLSH HDINMNHMDW DPSTANYVLP ETNMNINFPE KHTNEIGWVS SQVGYVPFEN 480
481 MIPSEVDINH MGMGYSGGSI PPQEETNTNN VGFVSCEIHS EIPPKTNMAI LETNYSNPLD 540
541 WVFPEDITSL ETNTIQKSLV SCETSYDALD NMVPLETNME EMNDALYDIS IEDLISFDID 600
601 ANEKDIFSWL EDNGFSEENN MMESCEYHNI ESVNQSDDMK IDDNFDDYRE CMDWINEEMN 660
661 KDV |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
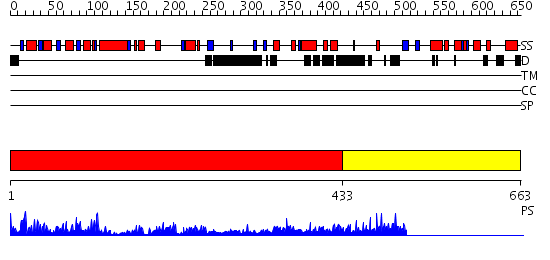
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..432] | 56.522879 | Crystal structure of sigm54 activator (AAA+ ATPase) in the inactive state | |
| 2 | View Details | [433..663] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||
| 1 |
|
||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)