













| General Information: |
|
| Name(s) found: |
gi|25403098
[NCBI NR]
gi|15220638 [NCBI NR] gi|12324175 [NCBI NR] |
| Description(s) found:
Found 12 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 711 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
RNA splicing
[IEA][ISS]
RNA-dependent DNA replication [IEA][ISS] |
| Molecular Function: |
RNA-directed DNA polymerase activity
[IEA][ISS]
RNA binding [IEA][ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKRLTYPLSS LRNIVEHFHC NTFHKPISSL SISPTLKSES REPSSTQDPY SLLKQDPVDI 60
61 CLSLWVKSFS SPPSATFSNL TGFLSKFDLW VLAYQRTCAH VTGTFPPRNA IHANALRSLL 120
121 SLQNAVTRSG GKFRWNDKMN QYVRSPKDKI SMNGGEGMSK GKVRRIIESE EPIFQDRVVH 180
181 EVLLMILEPF FEARFSSKSH GFRPGRNPHT VIRTIRSNFA GYLWFMKGDV SEMLDHVDVD 240
241 VVMNCLQKVV KDRKVLGLIE SSLKFSDKRV LKRVVEKHGN DNGLGTKRRI EREKRNKTKK 300
301 KILSDDEPKP DPYWLRTFYS FAPKEAAKVP SYGYCGVLSP LLANVCLNEL DRFMETKIVE 360
361 YFSPCKDDSI WKESIEDGCH NPAWPEFVPS SGKEKTRKMD YIRYGGHFLI GIRGPREDAV 420
421 KMRKEIIDFC DRVFGVRLDN SKLEIEHISR GIQFLDHIIC RRVIYPTLRY TGSGGSIVSK 480
481 KGVGTLLSVS ASLEQCIRQF RRLAFVKGDK DPEPLPCNPM LYSSQSHSNS QMNKFLETMA 540
541 DWYKYADNRK KAVGFCAYVI RSSLAKLYAA RYRLKSRAKV YSIASRDLSH PLSESSNNSA 600
601 PEYSDLLRMG LVDAIEGVQF SRMSLIPSCD YTPFPRNWIP NHEQVLQEYI RLQDPKFFCG 660
661 LHRSIKREGL TLPQDEISEA VWDFKTLGAW RSKYENKREA DDGLQKLDSQ T |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
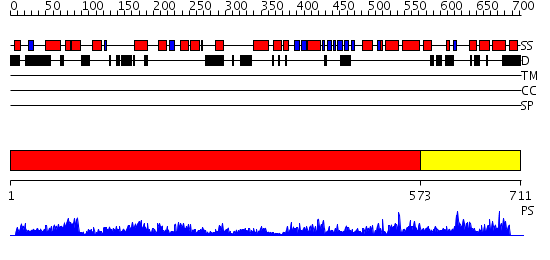
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..572] | 2.11 | Crystal structure of RNA-dependent RNA Polymerase 3D from human rhinovirus serotype 16 | |
| 2 | View Details | [573..711] | 3.134991 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||
| 1 |
|
||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)