













| General Information: |
|
| Name(s) found: |
ODPA2_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 16 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 393 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
mitochondrion [IDA] cytosol [IDA] |
| Biological Process: |
response to salt stress
[IEP]
metabolic process [ISS] |
| Molecular Function: |
zinc ion binding
[IDA]
pyruvate dehydrogenase (acetyl-transferring) activity [ISS] cobalt ion binding [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MALSRLSSRS NTFLKPAITA LPSSIRRHVS TDSSPITIET AVPFTSHLCE SPSRSVETSS 60
61 EEILAFFRDM ARMRRMEIAA DSLYKAKLIR GFCHLYDGQE ALAVGMEAAI TKKDAIITSY 120
121 RDHCTFIGRG GKLVDAFSEL MGRKTGCSHG KGGSMHFYKK DASFYGGHGI VGAQIPLGCG 180
181 LAFAQKYNKD EAVTFALYGD GAANQGQLFE ALNISALWDL PAILVCENNH YGMGTATWRS 240
241 AKSPAYFKRG DYVPGLKVDG MDALAVKQAC KFAKEHALKN GPIILEMDTY RYHGHSMSDP 300
301 GSTYRTRDEI SGVRQVRDPI ERVRKLLLTH DIATEKELKD MEKEIRKEVD DAVAQAKESP 360
361 IPDASELFTN MYVKDCGVES FGADRKELKV TLP |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
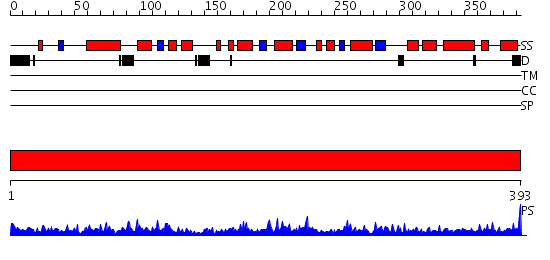
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..393] | 93.69897 | E1-beta subunit of pyruvate dehydrogenase (PP module) |
Functions predicted (by domain):
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.58 |
Source: Reynolds et al. (2008)