













| General Information: |
|
| Name(s) found: |
TCPD_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 14 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 536 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
response to cadmium ion
[IEP]
|
| Molecular Function: |
ATP binding
[IEA]
protein binding [IEA] unfolded protein binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAAVAAPMAS KPRGSKAESF VDNKRREDIR FANINSARAV SDAVRTSLGP KGMDKMISTA 60
61 NGEVIITNDG ATILNKMEVL QPAAKMLVEL SKSQDSAAGD GTTTVVVIAG ALLKECQSLL 120
121 TNGIHPTVIS DSLHKACGKA IDILTAMAVP VELTDRDSLV KSASTSLNSK VVSQYSTLLA 180
181 PLAVDAVLSV IDPEKPEIVD LRDIKIVKKL GGTVDDTHTV KGLVFDKKVS RAAGGPTRVE 240
241 NAKIAVIQFQ ISPPKTDIEQ SIVVSDYTQM DRILKEERNY ILGMIKKIKA TGCNVLLIQK 300
301 SILRDAVTDL SLHYLAKAKI MVIKDVERDE IEFVTKTLNC LPIANIEHFR AEKLGHADLV 360
361 EEASLGDGKI LKITGIKDMG RTTSVLVRGS NQLVLDEAER SLHDALCVVR CLVSKRFLIA 420
421 GGGAPEIELS RQLGAWAKVL HGMEGYCVKS FAEALEVIPY TLAENAGLNP IAIVTELRNK 480
481 HAQGEINAGI NVRKGQITNI LEENVVQPLL VSTSAITLAT ECVRMILKID DIVTVR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
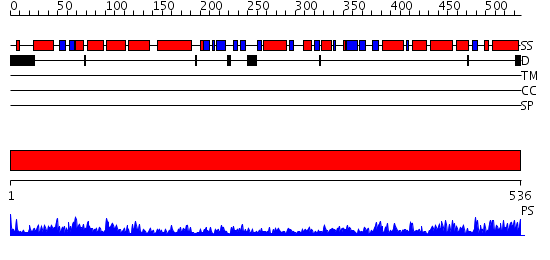
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..536] | 135.0 | Crystal structure of the chaperonin from Thermococcus strain KS-1 (nucleotide-free form of single mutant) |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)