













| General Information: |
|
| Name(s) found: |
GPDL6_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 14 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 753 amino acids |
Gene Ontology: |
|
| Cellular Component: |
anchored to membrane
[TAS]
|
| Biological Process: |
glycerol metabolic process
[IEA][ISS]
lipid metabolic process [IEA] |
| Molecular Function: |
glycerophosphodiester phosphodiesterase activity
[ISS]
kinase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLRFFILFSL FLHSSVAAPK TPAAAAAVPA KKWLTLNGQE PAVVARGGFS GLFPESSISA 60
61 NDLAIGTSSP GFTMLCNLQM TKDGVGLCLS DIRLDNATTI SSVFPKAQKT YKVNGQDLKG 120
121 WFVIDYDADT IFNKVTLVQN IFSRPSIFDG QMSVSAVEDV LGTKPPKFWL SVQYDAFYME 180
181 HKLSPAEYLR SLRFRGINVI SSPEIGFLKS IGMDAGRAKT KLIFEFKDPE AVEPTTNKKY 240
241 SEIQQNLAAI KAFASGVLVP KDYIWPIDSA KYLKPATTFV ADAHKAGLEV YASGFANDLR 300
301 TSFNYSYDPS AEYLQFVDNG QFSVDGVITD FPPTASQSIT CFSHQNGNLP KAGHALVITH 360
361 NGASGDYPGC TDLAYQKAID DGADIIDCSV QMSKDGIAFC HDAADLSAST TARTTFMSRA 420
421 TSVPEIQPTN GIFSFDLTWA EIQSVKPQIE NPFTATGFQR NPANKNAGKF TTLADFLELG 480
481 KAKAVTGVLI NIQNAAYLAS KKGLGVVDVV KSALTNSTLD KQSTQKVLIQ SDDSSVLSSF 540
541 EAVPPYTRVL SIDKEIGDAP KTSIEEIKKH ADAVNLLRTS LITVSQSFAT GKTNVVEEMH 600
601 KANISVYVSV LRNEYIAIAF DYFSDPTIEL ATFIAGRGVD GVITEFPATA TRYLRSPCSD 660
661 LNKDQPYAIL PADAGALLTV ADKEAQLPAI PPNPPLDAKD VIDPPLPPVA KLASNGTEGG 720
721 PPQTPPRSGT VAIAANLSLS LLAMMALGLL YTA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
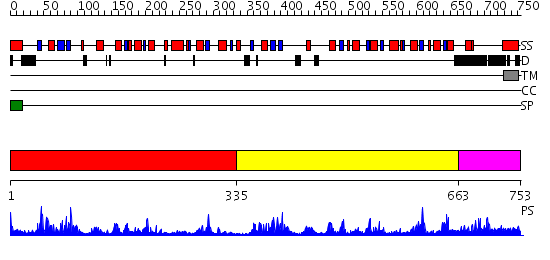
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..334] | 7.43 | Crystal structure of periplasmic glycerophosphodiester phosphodiesterase from Escherichia coli | |
| 2 | View Details | [335..662] | 62.0 | Crystal structure of periplasmic glycerophosphodiester phosphodiesterase from Escherichia coli | |
| 3 | View Details | [663..753] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|