













| General Information: |
|
| Name(s) found: |
MAOP1_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 581 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
malate metabolic process
[IEA][ISS]
metabolic process [IEA] oxidation reduction [IEA] |
| Molecular Function: |
malic enzyme activity
[ISS]
malate dehydrogenase (oxaloacetate-decarboxylating) (NADP+) activity [IDA] oxidoreductase activity, acting on NADH or NADPH, NAD or NADP as acceptor [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEKVTNSDLK SSVDGGVVDV YGEDSATIEH NITPWSLSVS SGYSLLRDPR YNKGLAFTEK 60
61 ERDTHYLRGL LPPVVLDQKL QEKRLLNNIR QYQFPLQKYM ALTELQERNE RLFYKLLIDN 120
121 VEELLPIVYT PTVGEACQKF GSIFRRPQGL FISLKDKGKI LDVLKNWPER NIQVIVVTDG 180
181 ERILGLGDLG CQGMGIPVGK LALYSALGGV RPSACLPVTI DVGTNNEKLL NDEFYIGLRQ 240
241 KRATGQEYSE LLNEFMSAVK QNYGEKVLIQ FEDFANHNAF ELLAKYSDTH LVFNDDIQGT 300
301 ASVVLAGLVS AQKLTNSPLA EHTFLFLGAG EAGTGIAELI ALYMSKQMNA SVEESRKKIW 360
361 LVDSKGLIVN SRKDSLQDFK KPWAHEHEPV KDLLGAIKAI KPTVLIGSSG VGRSFTKEVI 420
421 EAMSSINERP LIMALSNPTT QSECTAEEAY TWSKGRAIFA SGSPFDPVEY EGKVFVSTQA 480
481 NNAYIFPGFG LGLVISGAIR VHDDMLLAAA EALAGQVSKE NYEKGMIYPS FSSIRKISAQ 540
541 IAANVATKAY ELGLAGRLPR PKDIVKCAES SMYSPTYRLY R |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
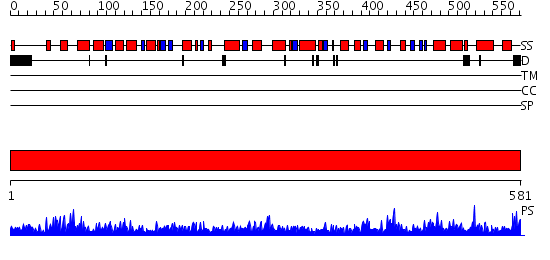
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..581] | 1000.0 | Mitochondrial NAD(P)-dependenent malic enzyme |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.80 |
Source: Reynolds et al. (2008)