













| General Information: |
|
| Name(s) found: |
EDS1C_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 623 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
aerenchyma formation
[IMP]
systemic acquired resistance [IEP] response to hypoxia [IMP] lipid metabolic process [ISS] systemic acquired resistance, salicylic acid mediated signaling pathway [IGI] response to singlet oxygen [IGI] defense response [ISS] plant-type hypersensitive response [IMP] regulation of hydrogen peroxide metabolic process [IMP] |
| Molecular Function: |
triglyceride lipase activity
[ISS]
lipase activity [ISS] signal transducer activity [TAS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAFEALTGIN GDLITRSWSA SKQAYLTERY HKEEAGAVVI FAFQPSFSEK DFFDPDNKSS 60
61 FGEIKLNRVQ FPCMRKIGKG DVATVNEAFL KNLEAIIDPR TSFQASVEMA VRSRKQIVFT 120
121 GHSSGGATAI LATVWYLEKY FIRNPNVYLE PRCVTFGAPL VGDSIFSHAL GREKWSRFFV 180
181 NFVSRFDIVP RIMLARKASV EETLPHVLAQ LDPRKSSVQE SEQRITEFYT RVMRDTSTVA 240
241 NQAVCELTGS AEAFLETLSS FLELSPYRPA GTFVFSTEKR LVAVNNSDAI LQMLFYTSQA 300
301 SDEQEWSLIP FRSIRDHHSY EELVQSMGKK LFNHLDGENS IESTLNDLGV STRGRQYVQA 360
361 ALEEEKKRVE NQKKIIQVIE QERFLKKLAW IEDEYKPKCQ AHKNGYYDSF KVSNEENDFK 420
421 ANVKRAELAG VFDEVLGLMK KCQLPDEFEG DIDWIKLATR YRRLVEPLDI ANYHRHLKNE 480
481 DTGPYMKRGR PTRYIYAQRG YEHYILKPNG MIAEDVFWNK VNGLNLGLQL EEIQETLKNS 540
541 GSECGSCFWA EVEELKGKPY EEVEVRVKTL EGMLGEWITD GEVDDKEIFL EGSTFRKWWI 600
601 TLPKNHKSHS PLRDYMMDEI TDT |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
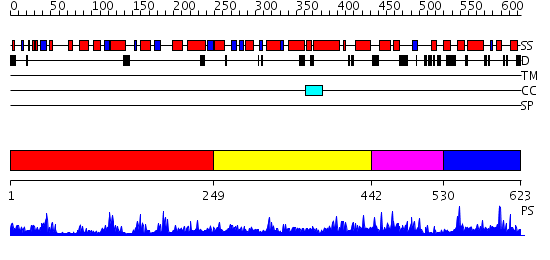
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..248] | 43.69897 | Crystal Structure of Ferulic Acid Esterase from Aspergillus niger | |
| 2 | View Details | [249..441] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [442..529] | 1.010967 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [530..623] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 |
|
|||||||||
| 2 | No functions predicted. | |||||||||
| 3 | No functions predicted. | |||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.55 |
Source: Reynolds et al. (2008)