













| General Information: |
|
| Name(s) found: |
ODP25_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 13 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 465 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast
[IDA]
cytosolic ribosome [IDA] chloroplast envelope [IDA] plasma membrane [IDA] |
| Biological Process: |
embryonic development ending in seed dormancy
[NAS]
|
| Molecular Function: |
protein binding
[IEA]
acyltransferase activity [IEA] dihydrolipoyllysine-residue acetyltransferase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSRLLQTPFL PSVSLPTKTR SSVTGFRVKP RIIPIQAKIR EIFMPALSST MTEGKIVSWV 60
61 KSEGDKLNKG ESVVVVESDK ADMDVETFYD GYLAAIMVEE GGVAPVGSAI ALLAETEDEI 120
121 ADAKAKASGG GGGGDSKAPP ASPPTAAVEA PVSVEKKVAA APVSIKAVAA SAVHPASEGG 180
181 KRIVASPYAK KLAKELKVEL AGLVGSGPMG RIVAKDVEAV AAGGGVQAAV AVKEVVAAPG 240
241 VELGSVVPFT TMQGAVSRNM VESLGVPTFR VGYTISTDAL DALYKKIKSK GVTMTALLAK 300
301 ATALALAKHP VVNSSCRDGN SFVYNSSINV AVAVAIDGGL ITPVLQNADK VDIYSLSRKW 360
361 KELVDKARAK QLQPQEYNTG TFTLSNLGMF GVDRFDAILP PGTGAIMAVG ASQPSVVATK 420
421 DGRIGMKNQM QVNVTADHRV IYGADLAQFL QTLASIIEDP KDLTF |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
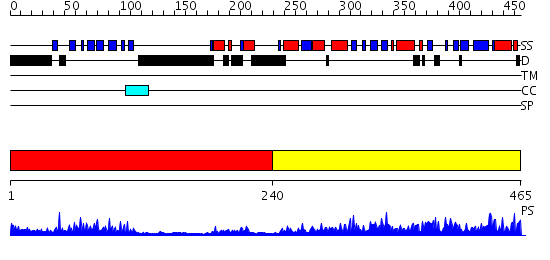
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..239] | 23.221849 | The crystal structure of dihydrolipoamide dehydrogenase and dihydrolipoamide dehydrogenase-binding protein (didomain) subcomplex of human pyruvate dehydrogenase complex. | |
| 2 | View Details | [240..465] | 83.39794 | Dihydrolipoamide succinyltransferase |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Source: Reynolds et al. 2008. Manuscript submitted |

|