













| General Information: |
|
| Name(s) found: |
MYST1_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 16 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 445 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
|
| Biological Process: |
chromatin assembly or disassembly
[IEA]
regulation of transcription [IEA] |
| Molecular Function: |
nucleic acid binding
[ISS]
histone acetyltransferase activity [IDA][ISS] zinc ion binding [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGSSADTETA MIIATPASNH NNPATNGGDA NQNHTSGAIL ALTNSESDAS KKRRMGVLPL 60
61 EVGTRVMCQW RDGKYHPVKV IERRKNYNGG HNDYEYYVHY TEFNRRLDEW IKLEQLDLDS 120
121 VECALDEKVE DKVTSLKMTR HQKRKIDETH VEGHEELDAA SLREHEEFTK VKNIATIELG 180
181 KYEIETWYFS PFPPEYNDCV KLFFCEFCLS FMKRKEQLQR HMRKCDLKHP PGDEIYRSST 240
241 LSMFEVDGKK NKVYAQNLCY LAKLFLDHKT LYYDVDLFLF YILCECDDRG CHMVGYFSKE 300
301 KHSEEAYNLA CILTLPPYQR KGYGKFLIAF SYELSKKEGK VGTPERPLSD LGLVSYRGYW 360
361 TRILLDILKK HKGNISIKEL SDMTAIKAED ILSTLQSLEL IQYRKGQHVI CADPKVLDRH 420
421 LKAAGRGGLD VDVSKMIWTP YKEQS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
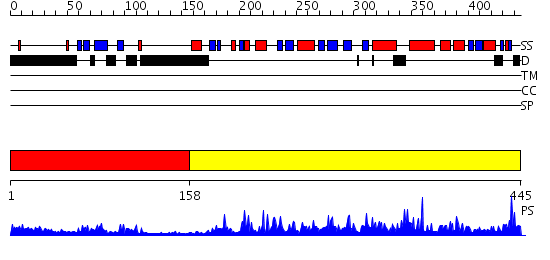
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..157] | 15.69897 | Solution Structure of the Tudor Domain from Mouse Hypothetical Protein Homologous to Histone Acetyltransferase | |
| 2 | View Details | [158..445] | 116.0 | No description for 2ou2A was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)