













| General Information: |
|
| Name(s) found: |
MAO1_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 623 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast
[IDA]
mitochondrion [IDA][ISS] |
| Biological Process: |
malate metabolic process
[ISS]
response to salt stress [IEP] |
| Molecular Function: |
ATP binding
[IDA]
zinc ion binding [IDA] malic enzyme activity [ISS] oxidoreductase activity, acting on NADH or NADPH, NAD or NADP as acceptor [ISS] cobalt ion binding [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGIANKLRLS SSSLSRILHR RILYSSAVRS FTTSEGHRPT IVHKQGLDIL HDPWFNKGTA 60
61 FTMTERNRLD LRGLLPPNVM DSEQQIFRFM TDLKRLEEQA RDGPSDPNAL AKWRILNRLH 120
121 DRNETMYYKV LINNIEEYAP IVYTPTVGLV CQNYSGLFRR PRGMYFSAED RGEMMSMVYN 180
181 WPAEQVDMIV VTDGSRILGL GDLGVHGIGI AVGKLDLYVA AAGINPQRVL PVMIDVGTNN 240
241 EKLRNDPMYL GLQQRRLEDD DYIDVIDEFM EAVYTRWPHV IVQFEDFQSK WAFKLLQRYR 300
301 CTYRMFNDDV QGTAGVAIAG LLGAVRAQGR PMIDFPKMKI VVAGAGSAGI GVLNAARKTM 360
361 ARMLGNTETA FDSAQSQFWV VDAQGLITEG RENIDPEAQP FARKTKEMER QGLKEGATLV 420
421 EVVREVKPDV LLGLSAVGGL FSKEVLEAMK GSTSTRPAIF AMSNPTKNAE CTPQDAFSIL 480
481 GENMIFASGS PFKNVEFGNG HVGHCNQGNN MYLFPGIGLG TLLSGAPIVS DGMLQAASEC 540
541 LAAYMSEEEV LEGIIYPPIS RIRDITKRIA AAVIKEAIEE DLVEGYREMD AREIQKLDEE 600
601 GLMEYVENNM WNPEYPTLVY KDD |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
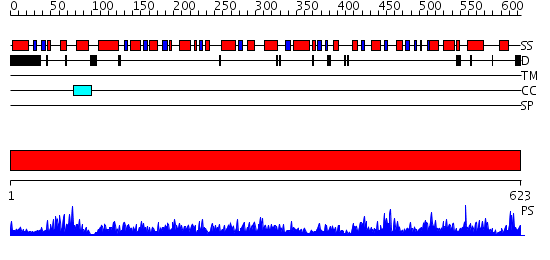
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..623] | 1000.0 | Mitochondrial NAD(P)-dependenent malic enzyme |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.67 |
Source: Reynolds et al. (2008)