













| General Information: |
|
| Name(s) found: |
CORI3_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 16 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 422 amino acids |
Gene Ontology: |
|
| Cellular Component: |
apoplast
[IDA]
vacuole [IDA] |
| Biological Process: |
hyperosmotic salinity response
[TAS]
response to abscisic acid stimulus [IEP] response to wounding [IEP] response to jasmonic acid stimulus [IEP] cellular amino acid metabolic process [IEP] biosynthetic process [ISS] response to microbial phytotoxin [IEP] |
| Molecular Function: |
transaminase activity
[ISS][NAS]
cystathionine beta-lyase activity [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MATLKCIDWQ FSGSEAAKDA AAASLGSYTS ALYALCDPHG KPILPPRNEI LETSNTAEKA 60
61 VVKAVLYGSG NAYAPSLGLA AAKSAVAEYL NQGLPKKLTA DDVFMTLGCK QAIELAVDIL 120
121 AKPKANVLLP SPGFPWDLVR SIYKNLEVRH YNFLPEKNFE IDFDSVRALV DENTFAIFII 180
181 NPHNPNGNTY SEAHLKQLAE LAKELKIMVV SDEVFRWTLF GSNPFVPMGK FSSIVPVVTL 240
241 GSISKGWKVP GWRTGWLTLH DLDGVFRNTK VLQAAQDFLQ INNNPPTVIQ AAIPDILEKT 300
301 PQEFFDKRQS FLKDKVEFGY SKLKYIPSLT CYMKPEACTF LWTELDLSSF VDIEDDQDFC 360
361 NKLAKEENLV VLPGIAFSQK NWLRHSIDME TPVLEDALER LKSFCDRHSN KKAPLKDVNG 420
421 VK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
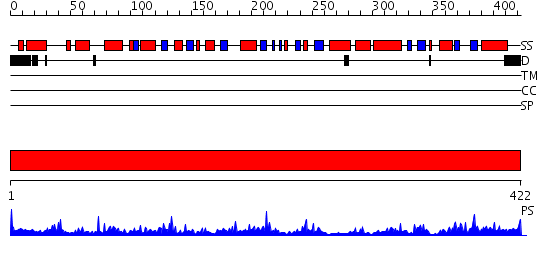
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..422] | 65.69897 | Tyrosine aminotransferase (TAT) |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)