













| General Information: |
|
| Name(s) found: |
gi|23296614
[NCBI NR]
gi|15241622 [NCBI NR] gi|13878039 [NCBI NR] gi|10178284 [NCBI NR] |
| Description(s) found:
Found 13 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 391 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
RNA binding
[IEA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAEVEEIAHE EQNIEKREKK KGKHEKPKPW DDDPNIDRWT IEKFDPAWNP TGMTETSTFS 60
61 TLFPQYREKY LQECWPRVES ALKEYGVACK LNLVEGSMTV STTRKTRDPY IIVKARDLIK 120
121 LLSRSVPAPQ AIKILEDEVQ CDIIKIGNLV RNKERFVKRR QRLVGPNSST LKALEILTNC 180
181 YILVQGSTVA AMGPFKGLKQ LRRIVEDCVQ NIMHPVYHIK TLMMKKELEK DPALANESWD 240
241 RFLPTFRKKN VKQKKPKSKE KKPYTPFPPP QPPSKIDMQL ESGEYFMSDK KKSEKKWQEK 300
301 QEKQSEKSTE NKRKRDASFL PPEEPMNNNS NANKSEDGKN DITELTNSLK SKTKELKKQK 360
361 KTHERVNAEE YIAGPSSSAD KSSKKSKKIR D |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
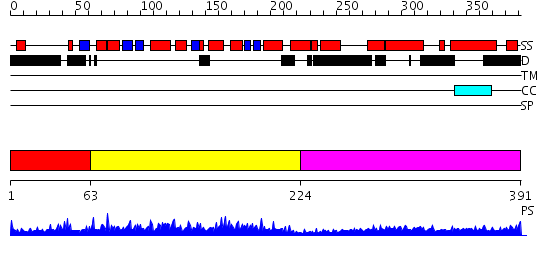
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..62] | 1.082988 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [63..223] | 29.154902 | Structural genomics, 1.5A crystal structure of a hypothetical protein APE0754 from Aeropyrum pernix | |
| 3 | View Details | [224..391] | 4.69897 | Heavy meromyosin subfragment |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)