













| General Information: |
|
| Name(s) found: |
DEAH8_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 11 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 883 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: | NONE FOUND |
| Molecular Function: |
nucleotide binding
[IEA]
nucleic acid binding [IEA] ATP binding [IEA] RNA helicase activity [ISS] nucleoside-triphosphatase activity [IEA] ATP-dependent helicase activity [IEA] helicase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 METNVPEKKI LGFPEEMNST IQFLSVEKTT CRLPVSIKFN IPKKISTKRR RISVEEEEEE 60
61 EDENKTKLSS WEKVLSSDEI LRLREVSRRK YLTDRENKKV EELRDERKDD DDVEGYRFPD 120
121 AYDQEGCIDQ KKRFDVAKER YCERRRSGRV VTEQEAWEDH QAQKARVRFG AKDKKQVVDG 180
181 YEFVFDDLTG FVEESSEAET GKHRGCYSKT AAEKAREGRE FLPIHGYREE LLKLIEENQV 240
241 LVIVGETGSG KTTQIPQYLQ EAGYTKRGKI GCTQPRRVAA MSVASRVAQE VGVKLGHEVG 300
301 YSIRFEDCTS EKTVIKYMTD GMLLRELLIE PKLDSYSVII IDEAHERTLS TDILFALVKD 360
361 VAKVRPDLRL IISSATLEAK KFSEYFDSAR IYLIPGRRYP VEKLFRKCPE PDYLETVIRT 420
421 VVQIHQTEAI GDILVFLTGQ EEIETVETNL KRRMMDLGTK GSEIIICPIY SNLPTPLQAK 480
481 VFEPAPKGTR KVVLATNIAE TSLTIDGVKY VIDPGYCKIN SYNPRTGMES LLVTPISKAS 540
541 AAQRAGRSGR TGPGKCFRLY NIKDLEPTTI PEIQRANLAS VVLTLKSLGI QDVFNFDFMD 600
601 PPPENALLKA LELLYALGAL DEIGEITKVG ERMVEFPVDP MLSKMIVGSE KYKCSKEIIT 660
661 IAAMLSVGNS VFYRPKNQQV FADKARMDFY EDTENVGDHI ALLRVYNSWK EENYSTQWCC 720
721 EKFIQSKSMK RARDIRDQLL GLLNKIGVEL TSNPNDLDAI KKAILAGFFP HSAKLQKNGS 780
781 YRRVKEPQTV YVHPNSGLFG ASPSKWLVYH ELVLTTKEYM RHTTEMKPEW LIEIAPHYYK 840
841 LKDIEDTRPK KTQRRIEEAS TSKVDTNKKT RTSKVDTNKK SKR |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
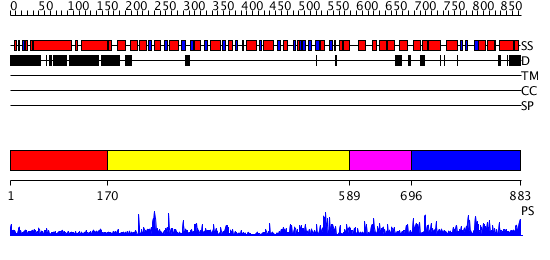
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..169] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [170..588] | 59.0 | No description for 2j0qA was found. | |
| 3 | View Details | [589..695] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [696..883] | 2.617986 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)