













| General Information: |
|
| Name(s) found: |
DAPB3_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 13 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 298 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast stroma
[IDA]
chloroplast [IDA] |
| Biological Process: |
lysine biosynthetic process via diaminopimelate
[ISS]
photosynthesis, light reaction [IMP] |
| Molecular Function: |
dihydrodipicolinate reductase activity
[IGI][ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAAVNCHFFQ LSRHLKPSRP SFSCSASQPS QNNIKVIING AAKEIGRAAV VAVTKARGME 60
61 LAGAVDNHFV GEDIGLLCDM EEPLEIPVVS DLTMVLGSIS QGKEVGVVID FTDPSTVYEN 120
121 VKQATAFGMK SVVYVPRIKP ETVSALSALC DKATMGCLVA PTLSIGSILL QQAVIMASFH 180
181 YNNVELVESR PNAADLPSPE AIQIANNISN LGQIYNREDS STDVQARGQV IGEDGVRVHS 240
241 MVLPGLPSST QVYFSSPGDV YTVKHDIIDV RSLMPGLLLA IRKVVRLKNL VYGLEKFL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
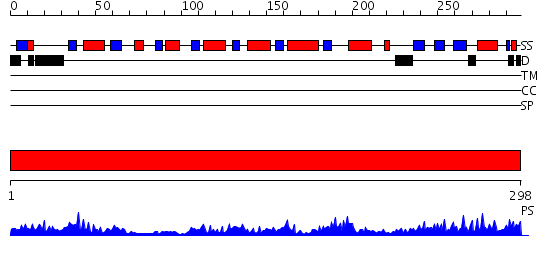
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..298] | 49.522879 | DIHYDRODIPICOLINATE REDUCTASE FROM MYCOBACTERIUM TUBERCULOSIS COMPLEXED WITH NADPH AND PDC |
Functions predicted (by domain):
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.89 |
Source: Reynolds et al. (2008)