













| General Information: |
|
| Name(s) found: |
CHR8_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 12 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 1187 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
DNA repair
[IMP]
response to gamma radiation [IMP] |
| Molecular Function: |
nucleic acid binding
[IEA]
ATP binding [IEA][ISS] DNA binding [IEA][ISS] helicase activity [IEA][ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEEDEDQFLL SSLGVTSANP EDLEQKILDE ATKKPDNDEG GSVEEKSTQL EGTNLLSSSQ 60
61 NELLNKLRAV KFEIDAVAST VENVDEIAAE KGLKKDDESD LQGLHSGSAL QHALATDRLR 120
121 SLKKRKIQLE KELTGLHGQS ASSSADHGNL LRDLVKEKPS LKRKLKEIRK PSRRDGKKVK 180
181 VVSFREDTDF DAVFDGASAG FVETERDELV RKGILTPFHK LDGFERRLQQ PGPSNSRNLP 240
241 EGDDENEDSS IIDRAVQSMS LAAKARPTTK LLDAEDLPKL EPPTAPFRRL RKLYKTPNSP 300
301 DNEAKKRKAG KKSKKTRPLP EKKWRKRISR EDSSLQGSGD GRRILTTSSC EEEELDDFDD 360
361 ADDNERSSVQ LEGGLNIPEC IFRKLFDYQR VGVQWLWELH CQRAGGIIGD EMGLGKTIQV 420
421 LSFLGSLHFS KMYKPSIIIC PVTLLRQWRR EAQKWYPDFH VEILHDSAQD SGHGKGQGKA 480
481 SESDYDSESS VDSDHEPKSK NTKKWDSLLN RVLNSESGLL ITTYEQLRLQ GEKLLNIEWG 540
541 YAVLDEGHRI RNPNSDITLV CKQLQTVHRI IMTGAPIQNK LTELWSLFDF VFPGKLGVLP 600
601 VFEAEFSVPI TVGGYANASP LQVSTAYRCA VVLRDLIMPY LLRRMKADVN AHLTKKTEHV 660
661 LFCSLTVEQR STYRAFLASS EVEQIFDGNR NSLYGIDVMR KICNHPDLLE REHSHQNPDY 720
721 GNPERSGKMK VVAEVLKVWK QQGHRVLLFS QTQQMLDILE SFLVANEYSY RRMDGLTPVK 780
781 QRMALIDEFN NSEDMFVFVL TTKVGGLGTN LTGANRVIIF DPDWNPSNDM QARERAWRIG 840
841 QKKDVTVYRL ITRGTIEEKV YHRQIYKHFL TNKILKNPQQ RRFFKARDMK DLFILKDDGD 900
901 SNASTETSNI FSQLAEEINI VGVQSDKKPE SDTQLALHKT AEGSSEQTDV EMTDKTGEAM 960
961 DEETNILKSL FDAHGIHSAV NHDAIMNAND EEEKMRLEHQ ASQVAQRAAE ALRQSRMLRS1020
1021 RESISVPTWT GRSGCAGAPS SVRRRFGSTV NSRLTQTGDK PSAIKNGISA GLSSGKAPSS1080
1081 AELLNRIRGS REQAIGVGLE QPQSSFPSSS GSSSRVGSLQ PEVLIRKICS FVQQKGGSAD1140
1141 TTSIVNHFRD IVSFNDKQLF KNLLKEIATL EKDQNRSFWV LKSEYKD |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
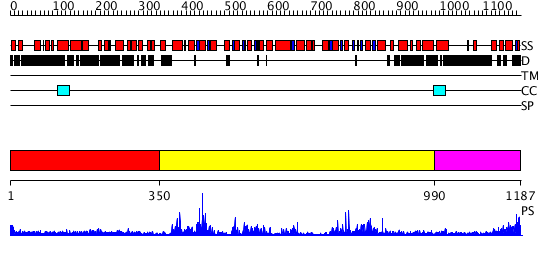
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..349] | 5.0 | Heavy meromyosin subfragment | |
| 2 | View Details | [350..989] | 94.154902 | Structure of the SWI2/SNF2 chromatin remodeling domain of eukaryotic Rad54 | |
| 3 | View Details | [990..1187] | 9.221849 | nucleosome recognition module of ISWI ATPase |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)