













| General Information: |
|
| Name(s) found: |
WUS_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 292 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA]
|
| Biological Process: |
maintenance of shoot apical meristem identity
[IGI]
stem cell maintenance [IMP] anther development [IMP] |
| Molecular Function: |
DNA binding
[ISS]
transcription factor activity [ISS][TAS] protein binding [IPI] transcription regulator activity [TAS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEPPQHQHHH HQADQESGNN NNNKSGSGGY TCRQTSTRWT PTTEQIKILK ELYYNNAIRS 60
61 PTADQIQKIT ARLRQFGKIE GKNVFYWFQN HKARERQKKR FNGTNMTTPS SSPNSVMMAA 120
121 NDHYHPLLHH HHGVPMQRPA NSVNVKLNQD HHLYHHNKPY PSFNNGNLNH ASSGTECGVV 180
181 NASNGYMSSH VYGSMEQDCS MNYNNVGGGW ANMDHHYSSA PYNFFDRAKP LFGLEGHQEE 240
241 EECGGDAYLE HRRTLPLFPM HGEDHINGGS GAIWKYGQSE VRPCASLELR LN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
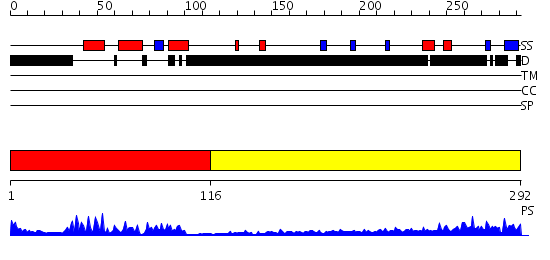
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..115] | 20.39794 | Crystal Structure of HoxA9 and Pbx1 homeodomains bound to DNA | |
| 2 | View Details | [116..292] | 8.659996 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 |
|
|||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)