













| General Information: |
|
| Name(s) found: |
CP41B_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 378 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast stroma
[IDA]
peroxisome [IDA] chloroplast [IDA] stromule [IDA] membrane [IDA] apoplast [IDA] chloroplast envelope [IDA] ribosome [IDA] plastoglobule [IDA] vacuole [IDA] |
| Biological Process: |
response to cold
[IEP]
rRNA processing [IGI] circadian rhythm [IEP][IMP] chloroplast organization [IMP] defense response to bacterium [IEP] |
| Molecular Function: |
coenzyme binding
[IEA]
catalytic activity [IEA] binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAKMMMLQQH QPSFSLLTSS LSDFNGAKLH LQVQYKRKVH QPKGALYVSA SSEKKILIMG 60
61 GTRFIGLFLS RILVKEGHQV TLFTRGKSPI AKQLPGESDQ DFADFSSKIL HLKGDRKDYD 120
121 FVKSSLSAEG FDVVYDINGR EAEEVEPILE ALPKLEQYIY CSSAGVYLKS DILPHCEEDA 180
181 VDPKSRHKGK LETESLLQSK GVNWTSIRPV YIYGPLNYNP VEEWFFHRLK AGRPIPVPNS 240
241 GIQISQLGHV KDLATAFLNV LGNEKASREI FNISGEKYVT FDGLAKACAK AGGFPEPEIV 300
301 HYNPKEFDFG KKKAFPFRDQ HFFASVEKAK HVLGWKPEFD LVEGLTDSYN LDFGRGTFRK 360
361 EADFTTDDMI LSKKLVLQ |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
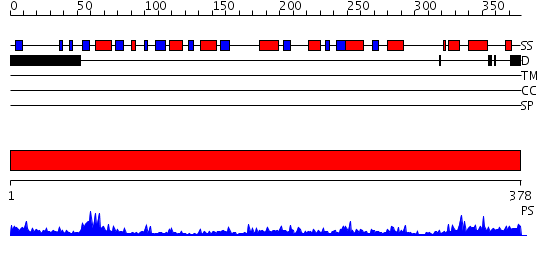
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..378] | 69.221849 | Crystal Structure of DesIV (dTDP-glucose 4,6-dehydratase) from Streptomyces venezuelae with NAD and TYD bound |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.53 |
Source: Reynolds et al. (2008)