













| General Information: |
|
| Name(s) found: |
ODP21_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 637 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrion
[IDA]
|
| Biological Process: |
metabolic process
[IEA]
pyruvate metabolic process [IEA] |
| Molecular Function: |
ATP binding
[IDA]
dihydrolipoyllysine-residue acetyltransferase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVLPLFRRAA IARTSSLLRA RLFAPASEFH SRFSNGLYHL DDKISSSNGV RSASIDLITR 60
61 MDDSSPKPIL RFGVQNFSST GPISQTVLAM PALSPTMSHG NVVKWMKKEG DKVEVGDVLC 120
121 EIETDKATVE FESQEEGFLA KILVTEGSKD IPVNEPIAIM VEEEDDIKNV PATIEGGRDG 180
181 KEETSAHQVM KPDESTQQKS SIQPDASDLP PHVVLEMPAL SPTMNQGNIA KWWKKEGDKI 240
241 EVGDVIGEIE TDKATLEFES LEEGYLAKIL IPEGSKDVAV GKPIALIVED AESIEAIKSS 300
301 SAGSSEVDTV KEVPDSVVDK PTERKAGFTK ISPAAKLLIL EHGLEASSIE ASGPYGTLLK 360
361 SDVVAAIASG KASKSSASTK KKQPSKETPS KSSSTSKPSV TQSDNNYEDF PNSQIRKIIA 420
421 KRLLESKQKI PHLYLQSDVV LDPLLAFRKE LQENHGVKVS VNDIVIKAVA VALRNVRQAN 480
481 AFWDAEKGDI VMCDSVDISI AVATEKGLMT PIIKNADQKS ISAISLEVKE LAQKARSGKL 540
541 APHEFQGGTF SISNLGMYPV DNFCAIINPP QAGILAVGRG NKVVEPVIGL DGIEKPSVVT 600
601 KMNVTLSADH RIFDGQVGAS FMSELRSNFE DVRRLLL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
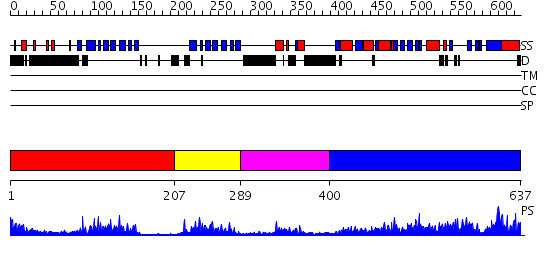
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..206] | 17.09691 | Crystal structure of the PDK3-L2 complex | |
| 2 | View Details | [207..288] | 12.30103 | No description for 2d5dA was found. | |
| 3 | View Details | [289..399] | 24.522879 | The crystal structure of dihydrolipoamide dehydrogenase and dihydrolipoamide dehydrogenase-binding protein (didomain) subcomplex of human pyruvate dehydrogenase complex. | |
| 4 | View Details | [400..637] | 81.69897 | Dihydrolipoamide succinyltransferase |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.57 |
Source: Reynolds et al. (2008)