













| General Information: |
|
| Name(s) found: |
AGD2_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 12 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 776 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IEA]
|
| Biological Process: |
regulation of ARF GTPase activity
[IEA]
|
| Molecular Function: |
zinc ion binding
[IEA]
protein binding [IEA] ARF GTPase activator activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAGFINLEDS PMFQKQVFSL EGTSDELKDR CQKLYKGVKK FMGALGEAST GVSAFADSLE 60
61 EFGAGHDDPV SVSIGGPVIS KFINTLRELS SYKEFLRSQV EHVLLERLTN FMTVDLQEAK 120
121 ESRRRFDKAV HSYDQAREKF VSLKKNTRGD IVAELEEDLE NSKSAFEKSR FNLVNSLMTI 180
181 EAKKKYEFLE SISAIMDSHF KYFKLGYDLL SQLEPYIHQV LTYAQQSKEQ SKIEQDRFAQ 240
241 RIQEFRTQSE LDSQQASAKA DPSDVGGNHV YRAIPRKNVE ANSVSTADKE VTKQGYLLKR 300
301 SASLRADWKR RFFVLDNHGS LYYYRNTGNK SAKSQHYYSG LGEHSSGVFG RFRTRHNRSA 360
361 SQGSLDCNMI DLRTSLIKLD AEDTDLRLCF RIISPQKTYT LQAENGADRM DWVNKITAAI 420
421 TIRLNSHFLQ QSPARYLDKK NTSSGPATEN LTLNQKEDYN QRLNVGDDVL TILREIPGNN 480
481 TCAECNAPDP DWASLNLGVL MCIECSGVHR NLGVHISKVR SLTLDVKVWE PTILDLFRNL 540
541 GNGYCNSVWE ELLHHLDDDS EKGSTDTLAS VSKPSSEDWF TLKEKYINGK YLEKALVVKD 600
601 EREANSTASS RIWEAVQSRN IRDIYRLIVK ADANIINTKF DDITDLDVYH HHHVDAPDEV 660
661 KKRHDPNACQ RIKNSNEARN CLQGCSLLHV ACQSGDPILL ELLLQFGADI NMRDYHGRTP 720
721 LHHCIASGNN AFAKVLLRRG ARPSIEDGGG LSVLERAMEM GAITDEELFL LLAECQ |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
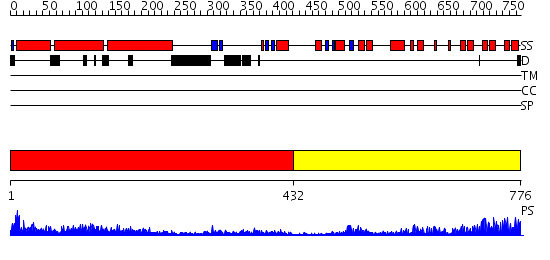
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..431] | 50.69897 | No description for 2elbA was found. | |
| 2 | View Details | [432..776] | 57.522879 | Ankyrin-R |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)