













| General Information: |
|
| Name(s) found: |
PI5K3_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 705 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA]
plasma membrane [IDA] apical plasma membrane [IDA] |
| Biological Process: |
root hair cell tip growth
[IMP]
root hair elongation [IMP] |
| Molecular Function: |
1-phosphatidylinositol-4-phosphate 5-kinase activity
[IDA][ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MQETVFLFTE ENLNKEQSLG VKYKQSSRRV VPMTSCEVSD TAAEIRIVEK VLKNGDLYNG 60
61 GLSAGVPHGT GKYLWSDGCM YEGEWTRGKA SGKGRFSWPS GATYEGQFKD GRMDGEGTFI 120
121 GIDGDTYRGH WLWGRKHGYG EKRYANGDGY QGNWKANLQD GNGRYVWSDG NEYVGEWKNG 180
181 VISGKGKMTW ANGNRYDGLW ENGAPVGKGV LSWGEEKTSY NGWGRKSKKK DEEIVQNHKL 240
241 SSVETLSANT NFPRICISEL EDTGVCDHVE ASPYTSESDT SGCGEQEWAR SPLLLESGGA 300
301 MSVQQSPRWL DEGDVKKPGH TVTAGHKNYD LMLNLQLGIR YSVGKHASLL RELRHSDFDP 360
361 KDKQWTRFPP EGSKSTPPHL SAEFKWKDYC PIVFRHLRDL FAIDQADYML AICGNESLRE 420
421 FASPGKSGSA FYLTQDERYM IKTMKKSEIK VLLKMLPNYY EHVSKYKNSL VTKFFGVHCV 480
481 KPVGGQKTRF IVMGNLFCSE YRIHKRFDLK GSSHGRTIDK DEGEIDETTT LKDLDLKYVF 540
541 RLETSWFQAF INQIDLDCEF LEAERIMDYS LLIGLHFRES GMRDDISLGI GRRDQEDKLM 600
601 RGNGPLMRLG ESTPAKAEQV SRFEEETWEE DAIDNSNPKG TRKEAVEVIL YFGVIDILQD 660
661 YDITKKLEHA YKSLHADPAS ISAVDPKLYS RRFRDFINKI FIEDK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
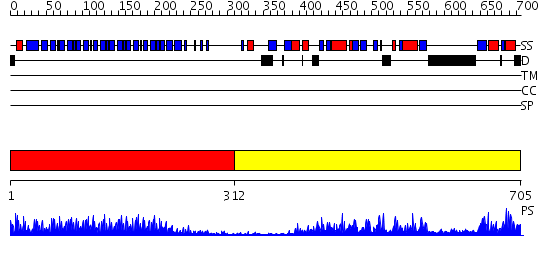
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..311] | 10.0 | Crystal structure of the complete modular teichioic acid phosphorylcholine esterase Pce (CbpE) from Streptococcus pneumoniae | |
| 2 | View Details | [312..705] | 91.30103 | Phosphatidylinositol phosphate kinase IIbeta, PIPK IIbeta |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)